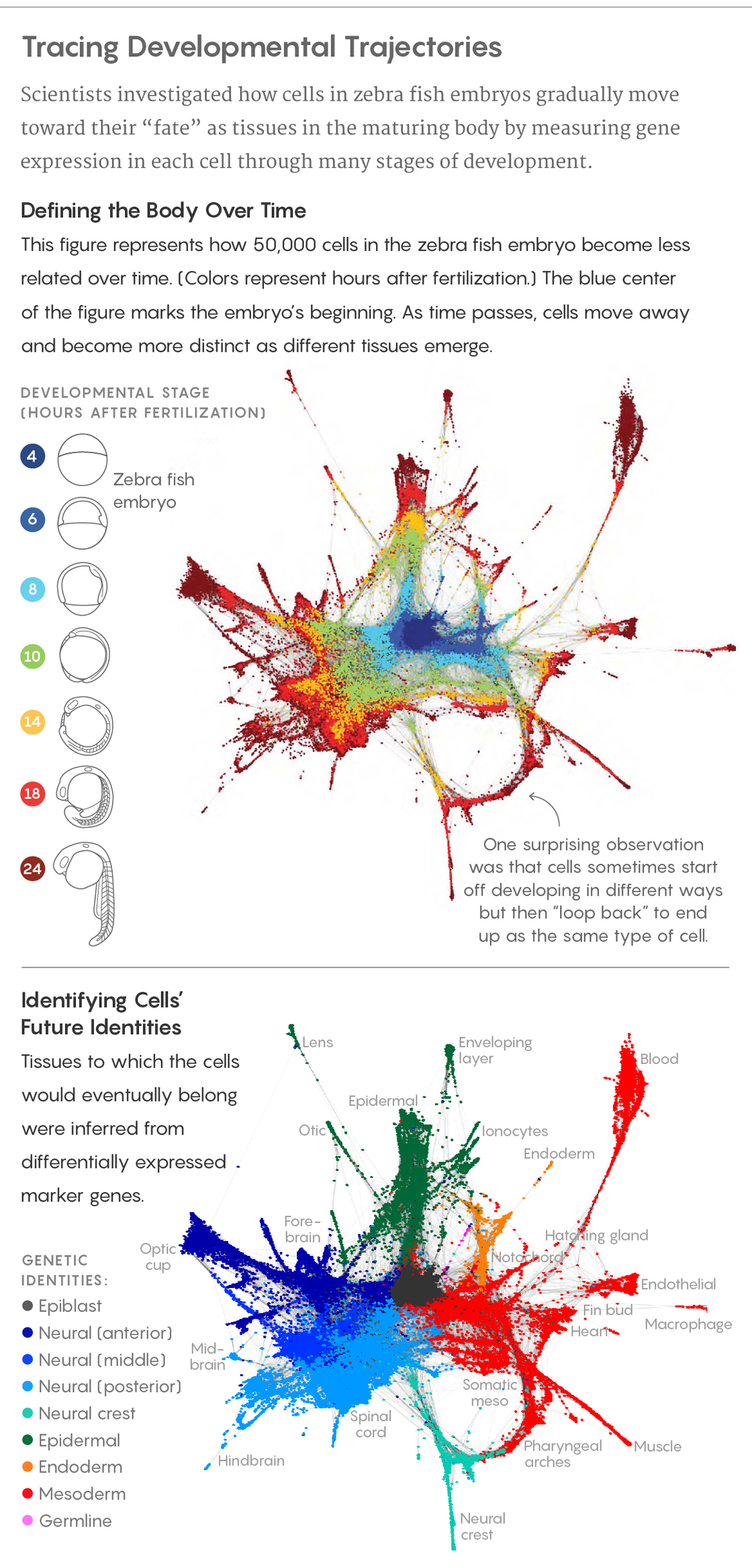
A fertilized egg divides first into two cells, then four, then eight and so on. Meanwhile, those cells progress from undifferentiated blobs in a cluster to more diverse identities associated with heart, brain, muscle, blood, bone and other tissues. Though the overall process is familiar, scientists have not understood it in much detail.
But three papers appearing today in Science are changing that, as they unveil work with major significance for the field of developmental biology. Using a combination of gene sequencing and mathematical methods, the researchers traced the patterns of gene expression in every cell in embryos of zebra fish and of Western clawed frogs through many stages of development during their first 24 hours.
The results revealed, at a previously impossible resolution and scale, the genetic and developmental trajectories that embryonic cells follow to their eventual fates in fully differentiated tissues. Surprising new insights emerged as well: Many biologists, for example, believed that embryonic cells always followed branching paths toward maturity that committed them irrevocably to certain fates. But the new data indicates that cells can, in effect, sometimes “loop back” to follow a different path, and that cells with different developmental histories can sometimes end up as the same type of cell.
The powerful techniques used in these reports, according to experts in the field, mark a new frontier in the ability to study development, cell fates and disease. “Whatever tissue you’re interested in studying, there’s something in this data set that should be of interest to you,” said Berthold Göttgens, a molecular biologist at the University of Cambridge who did not participate in the research but has been doing similar work in mouse embryos. Just as the rise of genome sequencing studies put biology on a different footing, he said, “this kind of foundational data will stand the test of time. It’ll be a landmark people will go back to.”
“There’s a whole universe of possibilities that data like this opens up,” said Alexander Schier, a cell biologist at Harvard University and an author on one of the studies. “Before, when we could only work with a few genes, or a few cells, or a few developmental stages, it was like we were seeing two or three stars. Now we can suddenly see an entire galaxy.”
Adapted from D. E. Wagner et al., Science 10.1126/science.aar4362 (2018)
Traditionally, developmental biologists have dyed cells in a dividing embryo and tracked their spatial trajectory, or they’ve targeted specific genes and examined their effects on the organism. More recently, they’ve also used editing techniques to integrate “bar codes” into cellular DNA; as cells divide, the bar codes acquire mutations that can be used to determine shared cellular lineages.
The research published in Science takes a different approach. In their two papers, the Harvard systems biologists Allon Klein, Marc Kirschner, Sean Megason and their colleagues measured the expression of messenger RNA in each of the cells; one paper combined that data with the bar-coding technology. The messenger RNA data defined each cell’s identity based on which genes it expressed, while the bar coding provided information on where that cell came from — its “family history,” so to speak.
Analysis of that data confirmed many findings reported by other scientists after years of painstaking investigation. “It was exciting to see decades of developmental biology research encoded in our data,” Klein said.
But their experiments also pushed ahead to new findings, too. “With the old techniques, you could imagine getting [the equivalent of] a map at the city level. But you still didn’t really know what made, say, Philly Philly compared to Pittsburgh,” said Leonard Zon, a stem cell biologist at Harvard Medical School who was not involved in the studies. “Now you get a description of that character: how it’s put together, where it came from, what it’s doing.”
The process of development is often visualized as something like a tree, with the fertilized egg at its base and later cells branching out and specializing until all the cell types have emerged. And that’s certainly what it looks like at first: Schier’s group, which studied zebra fish embryos over their first 12 hours, confirmed this topology.
But at later stages, “thinking of development as a tree can be a poor description of what’s going on,” Klein said. In defiance of old assumptions, branches sometimes come together, converging onto the same path in a loop. Cells with very different developmental histories can end up reaching very similar destinations.
In short, cells may be more plastic than scientists thought, and may truly commit to becoming specific tissues relatively late. “It’s another way to think about how to make tissues,” Zon said.
The finding, Klein added, taps into a larger question about what’s really happening during the process of development, and the significance of all those intermediate states for cells that don’t end up in the adult organism. If the sole purpose of development were to create specific cell types, then theoretically, the needed genetic programs could be switched on in the cells right away: Mature muscles, bones, nerves and other tissues could be present in the embryos from almost the very beginning. But instead, development is a process, and the right cells need to emerge at the right times and places to lay down patterns for growth and transformation in the embryo. Cells in transitional states may have unique functions and developmental value in themselves, and not just as steps toward some end. Different parts of the embryo require different intermediate steps, even if they’re ultimately going to give rise to the same cell type.