Main
In 1752, King Frederik V of Denmark, known for his “generous attitude […] towards natural science and applied art” commissioned the Flora Danica project, initiating an ‘Opus Incomparibile’ that took 122 years and produced one of the world’s most unique works in natural history6. Over 3,000 botanic engravings and 54 booklets were completed of flowers and plants, with which, “according to the unanimous contention of all connoisseurs”, “the whole world can eventually reap all the fruits that follow the extension of a science which, with regard to the benefit of mankind, is one of the most useful and without which medicine and economics would lack important advantages”6. In 2019, we initiated the Microflora Danica (MFD) project with the aim of cataloguing the microbiome of Denmark, in the hope that the microflora of Denmark can be similarly studied, and their riches contribute to the extension of science.
The MFD dataset
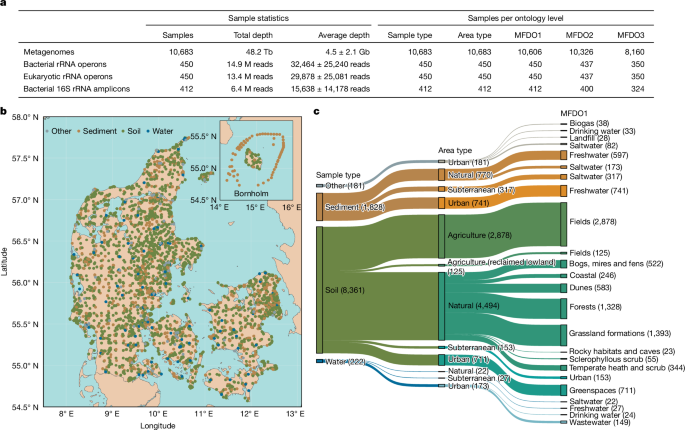
The MFD dataset comprises 10,683 samples, chosen to capture the diversity and geographical coverage of Danish microorganisms, associated with Illumina shotgun metagenomic DNA sequencing (average 4.5 Gb per sample, total 48.2 Tb) (Fig. 1a). Moreover, the dataset incorporates 14.9 million bacterial (median 4,528 bp) and 13.4 million eukaryotic rRNA operon sequences (median, 4,035 bp), as well as 6.4 million nearly full-length bacterial 16S rRNA gene sequences (median 1,355 bp). These data originate from a subset of samples (450 and 412, respectively) reflecting sample diversity while maintaining geographical coverage of the wider dataset (Fig. 1a). The samples are associated with GPS coordinates (Fig. 1b) and a highly curated five-level ontology (MFDO) (that is, habitat classification system) (Fig. 1c) that can be linked to other ontologies (EMPO3,7, Natura 2000 (ref. 8) and EUNIS9). The habitat ontology comprises sample type, area type and up to three levels of increasingly specific habitat description: MFDO1, MFDO2 and MFDO3 (MFD ontology levels 1, 2 and 3) (Fig. 1a,c). The area type ‘natural’ describes habitats not directly managed or located in urban areas. The Danish landscape is made up mainly of agriculture (63.0%), buildings and infrastructure (13.9%), forest (13.3%) and natural areas (9.2%), as well as streams and lakes (2.8%)10. The MFDO1 habitat ontology level represents 28 different categories (Fig. 1c) and reflects the primary land uses of fields (that is, croplands; 3,003 samples), grassland formations (1,393 samples), forests (1,328 samples) and greenspaces (that is, urban parks; 711 samples). The breadth of sampling is exemplified by our coverage of 27% of the 986 registered lakes in Denmark. Combined, the datasets, ontology, associated metadata and spatial resolution provide an extraordinary resource to investigate research questions related to diversity and function in microbial ecology.
a, The mean ± s.d. metagenome and rRNA amplicon data sequencing depths. The unit of measurement for depth is reads, except for metagenomes, for which the depth is reported as bp. M, million. b, The MFD samples cover the land of Denmark and its surrounding waters. The map depicts the locations of the samples used for metagenomics, and the colours represent the three different sample types. The top right cutouts show the island of Bornholm, which is east of Copenhagen and south of Sweden. The base map was retrieved from the Eurostat countries portal EuroGeographics for the administrative boundaries, © EuroGeographics 2025. c, Sample counts in the first three levels of the habitat ontology. The MFD habitat ontology accounts for a variable number of samples per category/branch. The Sankey diagram reports the first three levels of the ontology, and the thickness of the branches is proportional to the number of samples in each category. Only classes with n > 20 samples and non-empty MFDO1 classification are reported. Each habitat category is followed by the number of samples for that category in parentheses. The Sankey plot, including all five levels of the ontology, is provided in high resolution at Zenodo (https://doi.org/10.5281/zenodo.17162544).
Establishing Denmark’s microbiome
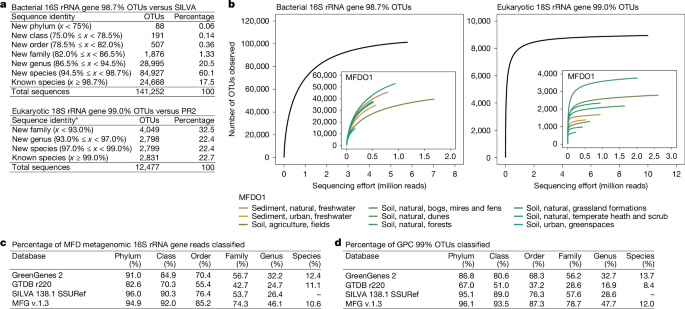
To facilitate sequence diversity analysis of bacteria, we used nearly full-length 16S rRNA genes extracted from the rRNA operon data, sequenced on the PacBio platform, and nearly full-length 16S rRNA gene amplicon data generated using unique molecular identifiers (UMIs). The UMI approach relies on the use of molecular nucleotide template tagging to achieve high-accuracy single-molecule consensus calling on the Oxford Nanopore sequencing platform. The addition of UMIs to both ends of the template enables the bioinformatic identification and removal of chimeras formed during PCR11. We investigated the bacterial sequence diversity and novelty using the combination of these data (Methods and Extended Data Fig. 1). The combined nearly full-length 16S (V1–V8) rRNA gene dataset included 458 habitat-representative samples and 21.3 million sequences, with 605,861 amplicon sequence variants (ASVs) representing 141,252 bacterial species (98.7% operational taxonomic units (OTUs))12 (Fig. 2a). Comparison of the species-level OTUs (clustered at 98.7% identity) against the SILVA v.138.1 database revealed that 82.5% were from new species (<98.7% identity) (Fig. 2a). However, the discovery rate of novelty quickly decreased at the higher taxonomic levels, with only 1.9% of the OTUs belonging to new families (<86.5% identity) (Fig. 2a). This suggests that while 16S rRNA gene sequences from bacteria originating from temperate northern European habitats are well represented in public databases at the higher taxonomic levels, the species-level diversity remains predominantly uncaptured.
a, Sequence novelty of species-level clustered bacterial 16S rRNA gene OTUs (98.7%) against SILVA19 v.138.1 NR99 and eukaryotic 18S rRNA gene OTUs (99.0%) against PR2 (ref. 16) v.5.0.0. Taxonomic thresholds for bacteria were adapted from ref. 12, whereas those for eukaryotes were calculated using a similar approach based on sequences from the PR2 v.5.0.0 database (Supplementary Note 1). Where indicated by an asterisk, thresholds were proposed on the basis of the sequence identity between species-level classified 18S rRNA gene sequences in the PR2 database and their closest relatives within and across ranks; meaningful thresholds above the family level could not be determined. b, Species-level rarefaction curves of UMI-based bacterial 16S rRNA and eukaryotic 18S rRNA gene OTUs from terrestrial samples. Insets: MFDO1 habitat-specific rarefaction curves for habitats represented by at least nine samples. c,d, Database evaluation based on 16S rRNA gene reads extracted from selected MFD metagenomes (c) and V4 OTUs clustered at 99% identity from the GPC23 dataset (d). Classification of metagenomic reads or OTUs was done using the SINTAX63 classifier. The following databases were used in addition to the MFG database created here: GreenGenes2_2022_10 taxonomy backbone22, GTDB_ssu_all_r220 (ref. 34 and SILVA_138.1_SSURef_NR99 (ref. 19). All databases were clustered at 98.7% sequence identity to enable direct comparison.
We used the nearly full-length 16S rRNA UMI dataset to estimate the Danish terrestrial bacterial richness (species count). The dataset encompasses 5.8 million 16S rRNA gene reads and 101,423 species (98.7% OTUs)12 across 309 habitat-representative samples (Fig. 2b). Rarefaction analysis showed underlying variation in the detection of species among MFDO1-level habitats, but approached saturation across the combined dataset, indicating that most species were captured by the sequencing effort (Fig. 2b). To support this, we calculated the habitat and pan-habitat community coverage to estimate how well our dataset captures the total terrestrial diversity of bacterial species13. We found that the community coverage at the MFDO1-level habitat ranged from 0.46 to 0.90, showing a strong correlation with sampling effort r7 = 0.95 (t = 7.96, 95% confidence interval (CI): 0.77–0.99, P = 9.4 × 10−5), but that the overall terrestrial community coverage amounted to 0.98, again indicating near complete species detection. Hill diversity estimates place the lower bound of the bacterial species count (Hill richness) in terrestrial MFD at a minimum of 114,400 species (95% CI = 113,897–114,902), with 43,447 common (intermediate to high frequency, Hill–Shannon14) and 22,036 dominant (most frequent, Hill–Simpson14) species based on their observation frequency in the dataset13,15. The community coverage estimates and rarefaction analysis indicate that the nearly full-length 16S rRNA gene dataset captures the collective Danish bacterial species pool across the investigated habitats and sets a conservative minimum estimate of the total environmental bacterial richness of terrestrial Denmark to be 114,400 species.
To investigate the diversity of eukaryotes, we used the eukaryotic rRNA operon sequences. These sequences exhibited a strong phylogenetic signal as they include both the ITS1 and ITS2 regions. However, the absence of a comprehensive rRNA operon reference database prompted us to focus our analysis on extracted nearly full-length (V4–V9) 18S rRNA genes that can be directly compared to the PR2 database16. The 13.4 million eukaryotic nearly full-length 18S rRNA gene sequences resolved into 28,575 ASVs representing 12,447 species (99% OTUs; Supplementary Note 1). Mapping of the species-representative sequences against PR2 revealed that most species (77%) are novel (Fig. 2a). Furthermore, 32% of the sequences had less than 93% similarity to a sequence in PR2, indicating high novelty at approximately the family level (Supplementary Note 1). Eukaryotic diversity varied between habitats but, based on Hill diversity estimates, the eukaryotic species count (Hill richness) is estimated to be a minimum of 19,295 species (Supplementary Note 2 and Extended Data Fig. 2). These findings show that vast microeukaryotic diversity remains undocumented.
MFG 16S rRNA gene database
Confident taxonomic assignment of 16S rRNA gene sequences relies on representative databases with clear taxonomic frameworks that include uncultured taxa17,18. As current universal reference databases lack the specificity we required, we used our extensive nearly full-length 16S rRNA gene dataset to create a comprehensive reference database for taxonomic classification of 16S rRNA gene reads extracted from our metagenomes. To enhance classification accuracy, we supplemented our sequences with high-quality sequences from SILVA v.138.1 SSURef NR99 (ref. 19), EMP500 (ref. 3), AGP70 (ref. 11), MiDAS20 and ref. 21 (Methods). This resulted in a total of 30.2 million sequences, which were processed using Autotax18 to create the Microflora Global (MFG) 16S rRNA gene reference database. The 1,034,840 unique ASVs were clustered at 98.7% nucleotide identity, representing 342,673 bacterial or archaeal species-level OTUs with a complete seven-level taxonomic string.
To evaluate the MFG 16S reference database, we first compared classification of metagenome-derived 16S rRNA gene fragments from a subset (n = 2,348; Methods) of our samples using both the MFG 16S reference database and publicly available databases clustered at the species level (98.7% identity) (Fig. 2c). We classified 46.1% (4.79 million out of 10.40 million) of all extracted 16S rRNA gene reads to genus level using the MFG 16S reference database, compared with 32.2% (3.35 million out of 10.40 million) classified by the second-best-performing database GreenGenes2 (ref. 22) (Fig. 2c). We next evaluated our database’s ability to classify data beyond Denmark’s temperate Northern Hemisphere habitats, using the Global Prokaryotic Census (GPC) V4 OTU dataset23 (Fig. 2d). The MFG 16S reference database was able to classify 47.7% (1.05 million out of 2.20 million) of the GPC OTUs at the genus level, compared with 32.7% (0.72 million out of 2.20 million) classified by GreenGenes2 (ref. 22). The combined results confirm that the MFG 16S reference database greatly improves classification not only for our samples, but for microbial profiling in general.
Diversity for habitat management
The level of microbial diversity in a habitat is often characterized by the alpha diversity, the richness in a single sample or average sample of a habitat and by the gamma diversity, the total observed richness of all samples within a habitat24. In contrast to the aboveground macro biodiversity, disturbed (that is, managed or directly affected by human activities) soils have been shown to have higher richness than undisturbed natural areas, both at continental and global scales24,25. Our detailed habitat ontology and the number of samples in each habitat type enabled us to re-evaluate these observations using both the metagenome-derived 16S rRNA gene fragments and the nearly full-length 16S UMI rRNA gene dataset taxonomically classified against the MFG 16S reference database.
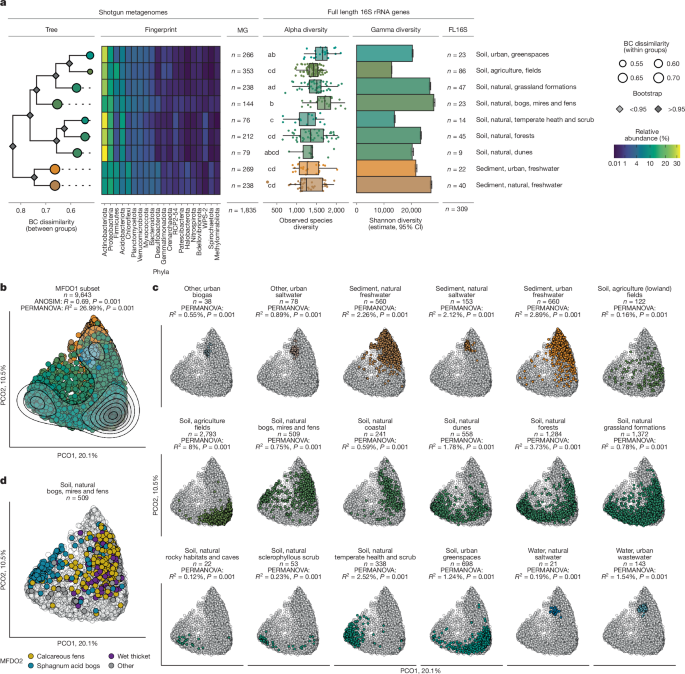
To ensure that our data enabled valid comparisons between samples, we investigated biases introduced from sample treatment and location. Most of the agricultural samples were treated differently compared with the other soil samples (Methods), but this treatment had no observable effect on alpha and beta diversity and amounted to only around 2% of the community variation (Supplementary Note 3). We accounted for spatial bias resulting from more densely sampled locations by estimating the spatial autocorrelation using distance–decay analysis on the metagenome-derived 16S rRNA gene fragments (Extended Data Fig. 3). On the basis of the results, we identified representative samples of the MFDO1 habitats within the 10 km reference grid of Denmark (2,348 samples; Methods and Extended Data Fig. 1). Hierarchical clustering based on the average between-habitat Bray–Curtis dissimilarity of these samples (beta diversity) largely captured the expected relationships based on similar aboveground characteristics (for example, grass cover, monocultures, exposure) between the habitats. These relationships are exemplified by the clustering of fields, greenspaces and grassland formations (Fig. 3a).
a, Diversity overview of the selected habitats. Each facet addresses a different measure of diversity. The nine MFDO1 habitats are represented in the rows of the multifacet plot. The dendrogram shows the between-group (branches) and within-group (nodes) Hellinger-transformed Bray–Curtis (BC) dissimilarity using the genus-level-classified 16S rRNA gene fragments from the spatially thinned dataset. Bootstrap values were calculated using 100 iterations. The heat map shows the relative abundances of the 20 most abundant phyla. The box plots of alpha diversity and bar charts of gamma diversity are based on the UMI 16S rRNA gene data. The number of biologically independent samples used for the diversity measurements per habitat are indicated. The hinges of the box plots correspond to 25th, 50th and 75th percentiles of the distributions, and the whiskers extend to 1.5× the distance between the 25th and 75th percentile. All individual samples are shown as points (with jitter for visualization). Gamma diversity (Hill–Shannon diversity) reflects a single value per habitat (that is, bar) based on rarefaction and extrapolation of n samples, and the error bars report the associated 95% CIs. b, Ordination of the metagenome (MG) dataset. PCoA of the 9,643 metagenome samples and coloured according to MFDO1 habitat description together with the results from the ANOSIM and PERMANOVA; P values were derived from 999 permutations in both cases. The visualization depicts the first two components. The contour plot was added to show the density of points. c, Subpanels of the individual 18 selected MFDO1 habitats coloured and presented with the results of the contrasts analysis. d, The sample distribution in the ordination space for MFDO1 ‘soil, natural, bogs, mires and fens’, coloured by classifications at the MFDO2 ontology level.
We calculated the alpha diversity from the nearly full-length UMI 16S rRNA gene data. In contrast to previous studies at the continental-Europe25 and global24 scale, which found the highest alpha diversity in samples from disturbed habitats, we found that the median bacterial diversity was highest in bogs, mires and fens (1,705 species) and lowest in temperate heath and scrub (1,274), with the diversity of disturbed habitats ranging in between (Fig. 3a and Supplementary Note 4). We found no significant difference in alpha diversity between fields, forests or grassland formations, contradicting the previous results from continental Europe while agreeing with global findings (Fig. 3a, Extended Data Table 1 and Supplementary Note 4). Additional large studies on other continents will be vital to resolving the effect of human disturbance on alpha diversity.
In contrast to the alpha diversity results, gamma diversity revealed key differences between disturbed and natural habitats (Supplementary Note 4). Fields had the lowest gamma diversity (12,797 common species), and along with greenspaces (20,336), was considerably less diverse than the more natural environment grassland formations (26,721). This trend was mirrored by sediments, with urban sediments (21,609) encompassing lower gamma diversity than natural sediments (27,126). Human disturbance reduces ecological breadth by creating more uniform environmental conditions, leading to lower gamma diversity26. This was supported by our comparison between urban and natural environments, in which greater environmental heterogeneity is encompassed by the natural habitats, reflecting greater habitat breadth and, consequently, higher gamma diversity. Overall, these data suggest that there is a gamma diversity gradient impacted by the level of perturbation, from highly disturbed fields to moderately disturbed greenspaces and relatively undisturbed grassland formations. These findings support an apparent homogenization (that is, low gamma diversity considering the high alpha diversity) of species in disturbed habitats—a pattern that was recently identified in other studies27. Habitat species homogenization was also supported by the Bray–Curtis analysis, which revealed low within-habitat dissimilarity of the prokaryotic communities (Fig. 3a).
Low gamma diversity in fields was most pronounced among bacterial communities (Fig. 3a), but also visible in the eukaryotic data (Supplementary Note 2), and reflected the aboveground macro biodiversity. Notably, temperate heath and scrub had similarly low gamma diversity to fields, but also low alpha diversity. However, this habitat is selective, defined by dry, infertile and acidic conditions (EUNIS habitat classification9), in contrast to the irrigated, nutrient- and pH-adjusted agricultural land.
These results show that the same bacterial species are found in the disturbed habitats, and that disturbed habitats are under selective pressures comparable to natural habitats with defined abiotic constraints. This highlights the need to incorporate gamma diversity when assessing microbial diversity. Including this broader perspective is particularly important when monitoring the impacts of land use and climate change, where community homogenization could lead to reduced ecosystem resilience and have implications for ecosystem functions28.
Modelling for habitat classification
After revealing the importance of gamma diversity for biodiversity assessments, we investigated how the microbial community could be used to classify habitats and its potential for tracking future habitat changes. Exploratory principal coordinates analysis (PCoA) performed on the eukaryotic 18S rRNA gene dataset revealed some separation between MFDO1 habitat categories (n = 363, analysis of similarities (ANOSIM), R = 0.46, P = 0.001, permutational analysis of variance (PERMANOVA), R2 = 0.07, P = 0.001; Supplementary Note 2). However, for the prokaryotic community, PCoA revealed good separation between MFDO1 habitats based on the metagenome-derived 16S rRNA gene fragment microbial community composition (n = 9,643, ANOSIM, R = 0.69, P = 0.001, PERMANOVA, R2 = 0.27, P = 0.001) (Fig. 3b,c). An exception was MFDO1 ‘bogs, mires and fens’, which showed large dispersion in ordination space. At the MFDO2 level, this habitat consists of both calcareous fens and sphagnum acid bogs, which have large differences in pH that impact microbial communities29 (Fig. 3d).
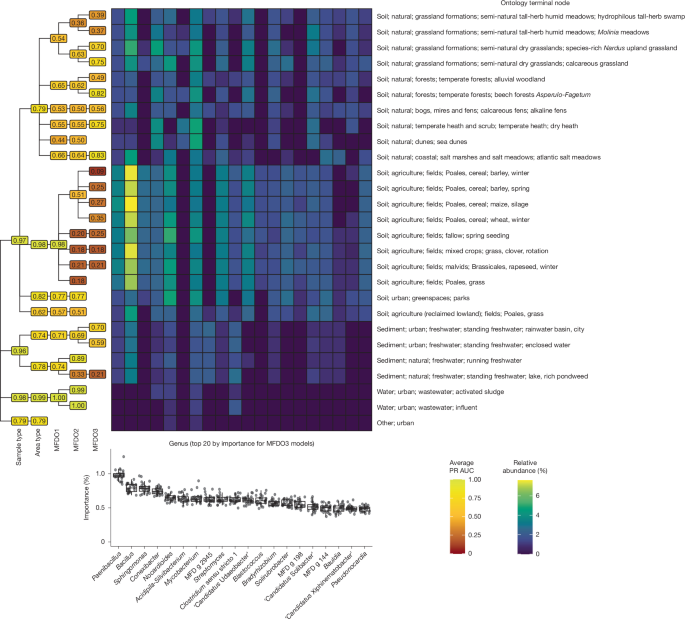
To determine the potential for microbial community DNA to be used in habitat classification, we investigated whether the 16S rRNA gene fragments could predict the habitat ontology (Fig. 4 and Supplementary Note 5). We evaluated habitat classifications using the precision recall area under the curve (PR-AUC; Fig. 4). Some habitats were difficult to model, for example, they had a lower PR-AUC (Fig. 4), such as the various types of fields, where the level of shared taxa was large. Conversely, other habitats—such as saltwater and wastewater, with higher PR-AUC—are associated with more specialized microbiomes. In general, low model scores reflected habitats in which samples would be misclassified to a few other selected habitats, for example, samples from grassland formations, greenspaces and fields were often misclassified as each other (Supplementary Note 5).
The genus-level models were used to compile the per-class PR-AUC of every node of the ontology. The metric spans from 0 to 1, where 0 and 1 mean that none and all, respectively, of the samples of the given class were classified correctly. The mean results over iterations (n = 25 independent iterations) are reported in the tree labels and coloured accordingly, with brighter nodes carrying higher values. Moreover, the top 20 genera, according to variable importance (box plot at the bottom, computed using the MFDO3 models), are reported with their median relative abundance for each of the terminal nodes of the ontology. The three hinges of the box plots correspond to the 25th, 50th and 75th percentiles of the distributions, and the whiskers extend to a maximum of 1.5× the distance between the 25th and 75th percentile hinges. All of the individual samples are shown as points (with jitter to improve visualization). The sum of the variable importance across all variables was scaled to 100 for each model. The ranking of the variables indicates which genera have a greater discriminant power in the models. Notably, the models were reliable in classifying samples from agricultural soils (PR-AUC =0.95) but not at classifying individual crop types.
Considering which prokaryotic genera were the most important in discriminating among habitats (that is, highest variable importance) (Fig. 4), the strongest signal was provided by Paenibacillus, whose species have been found to be associated with crops, promoting plant growth and protection from pathogens, as well as fixation of nitrogen30. Paenibacillus was distributed across soils and sediments with higher counts in field habitats, perhaps functioning as a predictor for sample type and land use. Our findings support low-resolution discrete habitat classification (that is, MFDO1) using microorganisms, but not higher-resolution classifications (that is, MFDO2). This agrees with previous studies proposing the redefinition of habitats using continuous gradients31. We believe microbiome data could provide a scalable solution to future classification efforts, enabling gradients to be compared to measure or monitor changes related to climate, sustainable farming choices or restoration progress. Identifying the core microorganisms belonging to specific habitats, or habitat gradients, may help to simplify the use of microbiome data.
Core genera across Danish habitats
Core microorganisms are abundant and widespread within habitats, potentially reflecting populations with habitat-specific adaptations, functions and ecological importance32. We identified abundant core community genera in the habitats across all five habitat ontology levels (genera with more than 50% habitat-specific prevalence, as well as at least 0.1% relative abundance; Supplementary Data 1, Extended Data Fig. 4 and Supplementary Note 6).
Habitat-specific core genera were more numerous in habitats with strong selective environmental gradients (for example, halotolerance), or constrained habitats, such as biogas systems (Supplementary Data 2 and Extended Data Fig. 4). Conversely, we observed fewer habitat-specific core species if no habitat-specific selective pressure was present. For example, the median of core genera unique to the soil MFDO1 habitats was two, showing that many of the genera were shared among two or more MFDO1 habitats (such as fields and greenspaces). Combined with the observed model misclassification of ecologically similar environments, these findings suggest that despite the vast dispersal capabilities of microorganisms, the prokaryotic community follows a continuous gradient of change and is thus more influenced by specific environmental factors as opposed to geographical location, in accordance with the Baas Becking hypothesis: everything is everywhere, but the environment selects33.
The alpha, beta and gamma diversity patterns, and high model score for fields among the terrestrial environments (Fig. 4), showed that land disturbance and management lead to similar microbial communities (Fig. 3a). Land-management practices, such as nutrient amendment and soil structure degradation, probably drive environmental filtering of the prokaryotic communities27. The disturbed soil habitats (fields, roadside and greenspaces) and the natural soil habitats (bogs, mires and fens; coastal; dunes; forests; grassland formations; rocky habitats and caves; sclerophyllous scrub; and temperate heath and scrub), encompassed 107 and 98 core genera (that is, a core genus in at least one of the habitats under the disturbed or natural categories), respectively (Supplementary Note 6). Comparing the natural and disturbed habitats revealed differences in core genera associated with nitrogen cycling (for example, Nitrospira, and genera within the Nitrososphaeraceae and Nitrosomonadaceae; Extended Data Fig. 4 and Supplementary Note 6), leading us to investigate this functional group more closely.
To provide genome-level resolution, recover potential functional group members and improve the representativeness of public genome databases, such as the Genome Taxonomy Database34 (GTDB), we performed de novo assembly of the 10,683 metagenomes (Supplementary Note 7). We recovered 19,253 bacterial and archaeal metagenome assembled genomes (MAGs) of at least medium quality (Methods and Extended Data Fig. 5). These MAGs represented 5,518 species (95% average nucleotide identity clustering) with broad phylogenetic coverage of which 4,604 were novel compared with GTDB34 R220 (Supplementary Note 7). This MFD genome database provides the foundation for functional analysis linked to species identity and habitat distribution and enabled us to examine key participants in the biogeochemical nitrogen cycle, the nitrifiers, across Denmark.
Distribution of Danish nitrifiers
Our investigations into microbial diversity indicated that bacteria and archaea involved in the nitrogen cycle were abundant, and form part of the core community differences in disturbed versus natural habitats (Supplementary Notes 6 and 8). This microbiome fingerprint reflects that Denmark is one of the most intensively cultivated countries in the world (63% of the land10), with much of its land impacted by management regimes involving fertilization with reactive nitrogen35. As Denmark has a large livestock sector, manure is a major nitrogen source, alongside synthetic fertilizers. Conversion of nitrogen fertilizers by nitrifying microorganisms leads to fertilizer loss, groundwater nitrate contamination, eutrophication of aquatic water bodies, and production of the potent ozone-depleting and greenhouse gas nitrous oxide35,36. Consequently, nitrification inhibition with synthetic or biological inhibitors is gaining importance to limit nitrate leaching, nitrous oxide emissions, and to increase nitrogen-use efficiency36. The use of two commercial nitrification inhibitors has risen fivefold in the past five years, now covering around 3% (78,129 ha in 2025) of Danish agricultural land37. Notably, the different groups of nitrifiers, comprising ammonia-oxidizing bacteria (AOB) and archaea (AOA), complete ammonia-oxidizing bacteria (CMX) and nitrite-oxidizing bacteria (NOB), vary in their sensitivities to nitrification inhibitors and in their nitrous oxide production rates36,38. To build knowledge needed to move towards sustainable agriculture, we performed an in-depth analysis of nitrifiers in the MFD datasets. On the basis of an analysis of functional genes (GraftM39), single-copy marker genes (SingleM40) and genome-level quantification (sylph41), we describe the diversity and distribution of Danish nitrifiers and identify new uncharacterized AOAs and NOBs.
Initially, curated gene-based search models of the nitrification marker genes amoA (encoding a subunit of the ammonia monooxygenase of AOB, AOA and CMX) and nxrA (encoding the active-site subunit of nitrite oxidoreductase of NOB and CMX) were created, accompanied by detailed classification of protein phylogeny from the translated genes, to separate nitrifier sequences from homologous sequences in other microorganisms, such as PmoA (particulate methane monooxygenase) and NarG (nitrate reductase)42. Furthermore, we included translated amoA and nxrA sequences from the recovered MFD MAGs in the search models, which markedly improved the resolution within groups of nitrifiers with few representative sequences (Fig. 5a and Supplementary Note 9).
a, Phylogenomic tree of nitrifiers. The red text indicates groups for which we recovered MAGs; the numbers in the brackets indicate the total number of species in this group in GTDB R220 (ref. 34), the number of species recovered in MFD, the number of species recovered in MFD not present in GTDB R220 and the total number of MAGs recovered in MFD. The values in parentheses represent the GTDB number of spp. representatives, the MFD number of spp. representatives, the MFD number of spp. representatives not in GTDB, and the MFD total number of MAGs, respectively. b, The distribution of nitrification genes across Danish habitats. The number of reads (reads per kilobase million (RPKM)) assigned to each gene-phylogenetic group (Supplementary Figs. 7 and 9). In cases in which a taxon has polyphyletic amoA or nxrA clades (Supplementary Note 9), an asterisk indicates the aggregation of multiple clades into a single line in the heat map. The samples are clustered with hierarchical clusters within each MFDO2 habitat. The bottom colour panel indicates the MFDO2 habitat. c, The distribution of canonical and potential nitrifiers across Danish habitats based on single-copy marker genes (SingleM). The heat map of short-read metagenomes is based on SingleM with the metapackage supplemented with MAGs from the MFD short-read metagenomes. Taxonomic resolution is marked by prefixes based on GTDB taxonomy: o__ (order), f__ (family) and g__ (genus). Reads assigned to higher taxonomic ranks, such as ‘f__Nitrososphaeraceae’, exclude descendants and consist only of those unassigned to a specific lower rank. pNxrA gene fragments assigned to the ‘Nitrospira_clade_1’ clade in the pNxrA tree (b) probably come from genomes of species in the GTDB genus Nitrospira_C (c), as the abundances of the pNxrA group and Nitrospira_C (based on single-copy marker genes) follow the same trends.
Analysis of the disturbed soil habitats showed similarities in their nitrifier communities, suggesting homogenization due to similar interferences, such as increased N availability, reduced aboveground diversity or physical soil disturbance. The highest relative gene abundances of canonical ammonia oxidizers (AOA and AOB) were observed in fields and greenspaces (Fig. 5b,c). These habitats were dominated by Nitrosospira AOB and Nitrososphaeraceae AOA. AOA distributions have been linked to soil acidity, as well as fertilization management regimes43. Furthermore, liming and inorganic fertilizer application in agricultural soils may create conditions in which Nitrosospira can also thrive43, as seen by the preference of Nitrosospira in fields. Similar to other studies44,45, AOA were more abundant than AOB in agricultural soils, in particular genera within the Nitrososphaeraceae, such as Nitrosocosmicus and several uncharacterized genera lacking isolates (TA-21, TH5893, TH5896, TH1177). Importantly, we were able to link these uncharacterized AOA genera to the major terrestrial amoA clades of uncharacterized groups through phylogenetic analysis of the amoA genes in their genomes (TA-21/NS-δ, TH5893/NS-γ 2.1, TH5896/NS-β 1 and TH1177/NS-ε)44,46 (Supplementary Data 3). By mapping nearly full-length 16S rRNA gene reference sequences to the MAGs, we linked two of these genera, TA-21 and TH5896, to core genera within the disturbed soil habitats (TA-21/MFD_g_198, TH5896/MFD_g_4907) (Supplementary Note 6).
Although AOA are generally abundant in agricultural soils, we identified a single undescribed AOA species, TA-21 sp02254895, that was highly abundant (sylph taxonomic abundance, median = 4.6%, maximum = 25.2%) across nearly all field samples and represented by 320 MFD MAGs (Fig. 5a, Extended Data Fig. 6 and Supplementary Note 8). Furthermore, the same species displayed lower relative abundance in agricultural field subhabitats (MFDO3) permanent grass, low yield (Mann–Whitney U-test, U = 10,628, one-sided, P = 1.23 × 10−5) and fallow fields, spring seeding (Mann–Whitney U-test, U = 41,104, one-sided, P = 0.045) (Fig. 5 and Supplementary Note 8) and was sparsely present in other non-agricultural soils except for urban parks (Mann–Whitney U-test, U = 90,748, one-sided, P = 3.01 × 10−20) and semi-natural grasslands (MFDO2), such as agricultural meadows (MFDO3) (Mann–Whitney U-test, U = 9,219, one-sided, P = 2.3 × 10−5) (Fig. 5, Extended Data Fig. 6 and Supplementary Note 8). The abundance of TA-21 sp02254895 across Denmark varied with land-use intensity and might be an effect of the level of anthropogenic disturbance. Owing to its link to disturbed Danish habitats, we propose the name ‘Candidatus Nitrososappho danica’.
Functional genome annotation of ‘Candidatus Nitrososappho danica’ revealed the potential to use ammonia (amoABC) and urea (ureABC) accompanied by ammonia (amt1/amt2) and urea transporters, CO2 fixation through the 3-hydroxypropionate/4-hydroxybutyrate cycle47 (acetyl-CoA/propionyl-CoA carboxylase (accC/pccC), methylmalonyl-CoA mutase (mcmA1, mcmA2), 4-hydroxybutyryl-CoA dehydratase (abfD)) and several genes involved in degradation of peptides (MEROPS IDs: cysteine C44, C26; serine S09C; threonine T01A; and metallopeptidases M38, M41, M48B) and polysaccharides (CAZyme IDs: GT2, GT55, GT81, CE1, CE14, CBM32, GH5). Specifically, GH5 and CBM32 were found in multiple copies (Supplementary Data 4), and have previously been reported to be highly expressed in the TA-21/NS-δ clade48. The mixotrophic potential of ‘Candidatus Nitrososappho danica’ might explain discrepancies between TA-21/NS-δ amoA abundance and nitrification and carbon-assimilation rates49, and future studies are needed to clarify whether this widespread and very abundant organism is growing through autotrophic ammonia oxidation. As N2O production is much lower from AOA than from AOB43,44,45, understanding the distribution and energy metabolism of archaeal species such as ‘Candidatus Nitrososappho danica’ will prove vital in managing the environmental impact of agricultural soils.
Moreover, recent studies suggest that CMX Nitrospira may be more abundant and important to soil nitrification processes than previously thought50. This is of particular interest, as CMX Nitrospira, like AOA, produce less N2O than canonical AOB do51. We identified Nitrospira as part of the core genera of disturbed soil habitats (Supplementary Note 6), but canonical and CMX Nitrospira are difficult to differentiate based on nxrA and 16S rRNA genes52. By using amoA gene phylogeny, we were able to assign CMX Nitrospira to its clade A and B subtypes52, which were found within the genera Nitrospira_D and Palsa-1315 (GTDB34 R220), respectively. Palsa-1315, first named from MAGs found in a permafrost peatland53, is probably a new comammox genus54. This is supported by a linear correlation (R2 = 0.48–0.81) between Nitrospira clade B amoA and Palsa-1315 nxrA in various MFDO1 habitats (Extended Data Fig. 7), and by identified amoA and nxrA in the recovered MAGs (Fig. 5a).
Notably, our improved search models showed that CMX clade B, for which no cultured representative is available, was more abundant than CMX clade A in most habitats, especially natural soils (Fig. 5b,c) and sediments (Supplementary Note 9). This challenges previous perceptions that CMX clade B is not abundant in forest soils55, wetland sediments56, and acidic or fertilized agricultural soils50,57. Nitrospira clade A amoA was inconsistently identified in sediments and agricultural soils, and was nearly absent from the other habitats investigated (Fig. 5b and Supplementary Note 9). Our analysis highlights CMX clade B as the most abundant ammonia oxidizer in Danish natural habitats, especially the MFDO2 habitats calcareous fens, alluvial woodland and semi-natural humid meadows, while canonical AOB and AOA were more abundant in disturbed habitats. Considering this, we propose the name ‘Candidatus Nitronatura plena’ for the species represented by a circular MAG, to describe the natural, widespread distribution of this most likely complete ammonia oxidizer. Nitrospira_C was the most abundant canonical NOB genus based on single-copy marker genes (Fig. 5c) and reads placed in the nxrA Nitrospira_clade_1 group (Fig. 5b), and displayed similar habitat patterns to the AOA, albeit at a lower abundance. At the national scale, it appears that nitrifier communities clearly reflect different habitat types, with their structure influenced by human impact. Here, we show that we can link specific species across land use types at scale.
Nitrobacter, a widely studied model organism of NOB, is considered abundant in fertilized soil based on nxrA identification58. However, our detailed search for NXR-encoding MAGs indicates that Nitrobacter may have been strongly overclassified, as we found NxrA sequences (>600 amino acids) phylogenetically falling between Nitrobacter and Nitrococcus (Extended Data Fig. 8 and Supplementary Notes 9 and 10). While other studies have reported cytoplasmic nxrA sequences clustering near, but outside of, cultivated Nitrobacter representatives in agricultural soils58, we were able to link these Nitrobacter-like NxrA sequences to Xanthobacteraceae family members, primarily Bradyrhizobium spp., Pseudolabrys spp. and the uncharacterized genus BOG-931 (Fig. 5 and Supplementary Note 10), which are not known to be NOB. In particular, a monophyletic clade of Nitrobacter-like NxrA from BOG-931 grouped closely to Nitrobacter and Nitrococcus NxrA, and the associated MAGs clustered together in a phylogenomic tree (Extended Data Fig. 8 and Supplementary Note 10).
We investigated gene synteny in long-read high-quality MAGs belonging to BOG-931 recovered from MFD samples59. Metabolic reconstruction revealed an operon resembling the nxr/nar operon of Nitrobacter winogradskyi and Nitrobacter hamburgensis60, consisting of cytochrome c class I, nxrA, nxrX, nxrB/narH, narJ and narI, and was flanked by transposases, accompanied by formate/nitrate transporters and cytochrome c oxidase gene clusters (Extended Data Fig. 9 and Supplementary Note 10). Consequently, Nxr-encoding members of BOG-931 could be potential new NOB occurring in many habitats, but confirmation requires culturing (Supplementary Note 10).
The putative Nitrobacter-like nxrA groups were found across fields, forests, grassland formations and greenspaces (Fig. 5b). In fields, Nitrobacter and Nitrobacter-like nxrA genes were present independent of crop type, but were less abundant than canonical Nitrospira NOB, such as Nitrospira_C (Fig. 5b,c). BOG-931 was most abundant in forest soils, and Nitrobacter and Nitrobacter-like NOB have previously been associated with nitrogen amendment in forest soils61. Indeed, BOG-931 was detected mainly in soil habitats lacking detected CMX clade B, and was particularly abundant in forests, grassland formations and sphagnum acid bogs of the bogs, mires and fens habitat (Fig. 5b). This suggests niche differentiation between CMX and canonical or potential NOB, and underlines a general need for further investigation of uncultured but abundant nitrifiers, including Nitrobacter nxrA-like containing groups, CMX clade B and AOA TA-21. As the presence and abundance of nitrifiers may be applied to evaluate how human activities affect the nitrogen cycle44, our results stress the importance of developing and applying reliable methods for quantitatively recording their diversity and distribution. Such methods must cover all important groups, including the newly detected nitrifiers, and provide insights into their response to environmental factors. The most critical of these factors is climate change, whereby the increased temperatures and longer growing seasons may lead to increased or prolonged nitrification activity, and more frequent droughts may lead to increased AOB activity but reduced AOA and CMX activity in soils62.
Conclusions
Here we provide an atlas of Denmark’s microbial communities, establishing a national baseline of microbial diversity. While many habitats have distinct microbial profiles, some show unexpected similarities undetectable through flora-inferred classification (for example, fields and greenspaces). These may result from land management disturbances, which enhance species diversity while also driving homogenization as communities affected by human disturbance converge. This homogenization extends to function, with nitrifier communities reflecting habitat type and human impact. Integrating gamma diversity metrics into biodiversity assessments may help to prevent national microbiome homogenization. Future assessments could adopt a data-driven approach, as our models show that short-read data can match microbiomes to flora-inferred habitats. The next step is linking microbial species and guilds, such as nitrifiers, to other national research efforts, including historical land use, fertilization regimes and greenhouse gas emissions. Through the identification and characterization of new species, microbially informed agricultural management is within reach, offering a potential strategy to limit N2O emissions by tailoring inputs to encourage or discourage specific microorganisms. We hope that other national atlases will follow, enabling comparisons of diversity and distribution on other continents. As we stand at the precipice of profound climatic shifts, the MFD dataset will be a vital resource for tracking microbial adaptations and resilience in both disturbed and natural ecosystems and a standard from which to monitor future restoration efforts.
Methods
Sampling
The MFD sample collection includes samples collected as part of the MFD sampling campaign, as well as samples contributed by members of the MFD consortium. The samples taken as part of the MFD sampling campaign were registered and associated with the appropriate metadata using codeREADr (https://www.codereadr.com) using a linear barcode attached to sterile 100 ml sample containers. After collection, the samples were stored between 4 °C and 10 °C for up to 48 h before being deposited at −20 °C for later processing.
As we wanted to cover as much of the Danish environmental landscape as possible, we requested expert collaborators send existing samples from interesting environments or environments that are not easily sampled. These include samples from existing publications, samples collected as part of governmental monitoring, but also samples from collaborators with no current publication. If not otherwise stated, these samples were acquired as frozen sample material. We divided each set of samples into projects, in which samples of the same type (soil, sediment, water) were subjected to the same treatment. Based on this, we constructed summary tables over the different protocols used for sampling and DNA extraction methodology (Supplementary Data 5). Most samples (across the biggest sample groups) were treated similarly, but we acknowledge that the different treatments might affect the results; consequently we applied the appropriate filtering where needed. The number of subsamples and other related sampling metadata are provided at GitHub and in Supplementary Data 6.
Soil samples
Topsoil samples from the MFD sampling campaign were collected as up to five subsamples (0–20 cm), taken within a ∼80 m2 (5 m radius) sampling area using a weed extractor, which was cleaned with 70% ethanol between sampling sites. As DNA from microorganisms could potentially be overwhelmed by the DNA from whole specimens in the sample material, we visually inspected each subsample with the naked eye and avoided including complete specimens (grass, leaves, sticks or larger animals) in the samples. After specimen removal on site, the subsamples were combined in a sterile plastic bag, the bag closed and the collective sample homogenized by hand before up to 100 ml was transferred to the barcoded sample container (P04_2, P04_4, P04_6, P04_7, P08_1, P08_2, P08_3, P08_5, P08_6, P08_7, P08_8 and P17_1). The samples from projects P19_1, P20_1, P21_1 (ref. 64) and P25_1 were collected as single subsamples. The subset of topsoil samples from the Land Use and Coverage Area Frame Survey (P04_8)25 were collected by collaborators from Aarhus University as described in previously65, in a manner very similar to the MFD sampling campaign.
A subset of the topsoil samples (P01_1)31 from both natural and agricultural habitats were provided by collaborators from Aarhus University and Copenhagen University. These were collected as described previously31. In brief, 81 subsamples, spanning a 9 × 9 grid covering a 40 × 40 m plot, were mixed into a representative sample from which we acquired a subsample. New sample projects were added to extend the existing project with wet terrestrial habitats (P01_2), agricultural and semi-agricultural habitats (P02_1), sites with different agricultural practices (P02_2 (ref. 66)) and urban habitats (P03_1 (ref. 67)). These were all collected as 81 subsamples except in the case of P03_1 which was mixed from 9 subsamples.
Samples from subterranean soils were collected as single samples from different depths using a soil drill (P06_1, P06_2, P06_3). Subsurface soil (P06_1) was collected with PVC liners by percussion hammering using a Geoprobe (NIRAS) drill rig. Soil samples were then collected around the oxic-anoxic interface with 5 ml cut-off syringes through openings cut into the core liners. We acquired the samples from P06_3 as DNA, which had previously been extracted using the DNeasy PowerLyzer PowerSoil Kit (QIAGEN) according to the manufacturer’s protocol. In the case of agricultural soils from croplands, six out of every seventh sample was provided by SEGES. As part of the collection, the individual samples were frozen, crushed to particles below 1 cm in size and dried at 37 °C (P04_3, P04_5), the effect of which was investigated (Supplementary Note 3).
Sediment samples
Surface sediment samples (0–10 cm) from the MFD sampling campaign were collected as up to five sediment subsamples from across the sampling area using a gravity corer, which was cleaned with 70% ethanol between sampling sites. The subsamples were combined in a sterile plastic bag, the bag was closed and the collective sample was homogenized by hand after careful removal of larger debris and any collected water. Up to 100 ml of the homogenized sample was transferred to the barcoded sample container. For the sediment from standing water sources (P05_1, P05_2, P08_5, P09_1, P09_2, P11_1 and P11_3) the top 10 cm was collected, while only the top 5 cm was collected from streams (P10_1, P10_2 and P10_3). Pond sediment (P09_2) was collected from the deepest point of each pond. Stream samples were collected as three subsamples across a 20 m transect of the stream, two at 25% distance from each brim and one in the middle of the stream.
Lake sediments provided by University of Southern Denmark were from either lakes selected for investigation of biotic phosphorus dynamics (P09_3) or a lake restauration initiative (P09_4). For P09_3, the cores were taken from the deepest part of the lake. For P09_4, the cores were taken at five different sampling stations. In both cases, a gravity corer was used for the sampling68. Sediment samples from coastal areas (P11_2) were collected at a single point using a HAPS bottom corer, as described previously69. Each sample was mixed from 10 subsamples of the sediment core (0–2 cm and 5–7 cm). We acquired the samples as DNA, which had previously been extracted with the DNeasy PowerMax Soil Kit (QIAGEN) according to the manufacturer’s protocol.
We acquired DNA extracts from Aarhus University from multiple sampling campaigns of marine surface and subterranean sediments (P12_1 (ref. 70), P12_2 (refs. 71,72)). P12_1 holds samples from the Bornholm Basin stations BB01 (13–63 cm) and BB03 (19–73 cm) sampled with a Rumohr corer. Samples from P12_1 were previously extracted with the DNeasy PowerLyzer PowerSoil Kit (QIAGEN) according to the manufacturer’s protocol while phenol-chloroform-isoamyl-alcohol extraction was used for P12_2. We expanded this sample category with sediments from the Baltic Sea provided by WSP Denmark (P12_5). Top sediments (0–30 cm) were collected using a HAPS bottom corer while subterranean sediments (0–300 cm) were collected using a Vibrocore sampler.
Water samples
Besides the samples from fjords (P11_3) the diversity in the natural environments found in the soil and sediment categories is not reflected in this category. The water category is instead made up of samples with a link to the urban environment: drinking water from the waterworks stage (P16_1 (ref. 73), P16_3 (ref. 74)), tap water (P16_1), potentially polluted groundwater (P16_4) and samples from wastewater treatment plants (P13_1 (ref. 75), P13_2 (ref. 75)).
Water samples from fjords (P11_3) were collected with a 1 l Ruttner water sampler. The sampler was rinsed three times with water from the locality before the water samples were collected. At each sampling location, five 1 l water samples were randomly collected. The water samples were transferred to 5 l cleansed plastic bottles and stored in a cooler (< 8 h) until they could be filtered in the laboratory. At stations with a halocline, water samples were collected from both above and below the halocline and were treated as two separate events. The collected water was filtered through a mixed cellulose ester membrane (47 mm, 0.22 µm) by dead-end filtration using a sterile filter funnel and a vacuum pump. The amount of water filtered for each sampling site varied from 0.3 l to 1 l. Filters were stored at −20 °C before DNA was extracted using the DNeasy PowerWater Kit (QIAGEN), according to the manufacturer’s protocol. Samples of drinking water (2 l) from the drinking water treatment plants (P16_1) were filtered through a mixed cellulose ester membrane (47 mm, 0.22 µm). The amount of water filtered varied from 0.25 l to 1.8 l. Tap water (5 l) was filtered through a cellulose acetate membrane (47 mm, 0.22 µm). The amount of water filtered varied from 4 l to 5 l. In both cases, filtering was performed as dead-end filtration using a sterile filter funnel and a vacuum pump and the filters were stored at −20 °C before DNA was extracted from the filters using the DNeasy PowerWater Kit (QIAGEN), according to the manufacturer’s protocol. The samples in P16_4 of probable polluted ground water (1 l) were filtered through a cellulose nitrate membrane (47 mm, 0.22 µm) by dead-end filtration using a sterile filter funnel and a vacuum pump. The amount of water filtered varied from 0.5 l to 1 l. Filters were stored at −20 °C before DNA was extracted from the filters using the DNeasy PowerLyzer Kit (QIAGEN) according to the manufacturer’s protocol.
The Technical University of Denmark provided DNA extracted from concentrates. Samples were taken from raw water (abstracted groundwater), filtered water (after secondary sand filters), treated water (after ultraviolet treatment) and water from the distribution network (P16_3). Between 100 l and 250 l of water from the different sampling points was pumped through separate REXEED 25S filters at a constant rate. After sample elution (200 ml) from the REXEED filter, a secondary concentration was conducted using VivaSpin 15R (SATORIUS) filters and centrifugation at 3,500g. DNA was extracted from 100 µl of final concentrate using the NucliSens miniMAG platform and NucliSens Magnetic Extraction Reagents (bioMerieux) according to the manufacturer’s protocol. Samples from wastewater treatment plants were included as DNA samples extracted with the FastDNA Spin Kit for Soil (MP Biomedicals). Wastewater was sampled as both influent (P13_1), using flow proportional sampling, and activated sludge from the aeration tank (P13_2) as described previously75.
Other samples
We included a last group of samples to encompass the samples that did not directly fit into either of the soil, sediment and water categories. This category covers samples from various surfaces in harbours (P18_1 and P18_2), sand filter material from drinking water treatment plants (P16_2 and P16_3), sludge from anaerobic digesters (P13_3), and scrapings from the walls of a limestone mine and a salt vat (P25_1). The harbour samples (P18_1 and P18_2) were collected as individual scraped-off biofilms and biocrusts from a range of different surfaces (such as fenders and piers). Each sampling location is associated with three individual samples. Sand filter material in P16_2 was collected as described previously76. The pooled medium samples were made by homogenizing and combining 20 g subsamples from 20 cm depth intervals. For P16_3 the sand filter material (around 15–40 ml) was collected from 1–2 locations at the top of 12 groundwater-fed rapid sand filters of 11 Danish waterworks using a 1% hypochlorite-wiped stainless-steel grab sampler. Samples from anaerobic digesters (P13_3) were collected from 20 digesters across Denmark, with DNA extracted using the standard DNeasy PowerSoil Pro Kit (QIAGEN) according to published protocols20.
Subsampling and DNA extraction
Unless otherwise stated in the precious sections, DNA extraction was performed as previously described77. The sample containers were thawed at 4 °C and dried soil samples were rehydrated using phosphate-buffered saline before subsampling. The sample material was divided into a total of three Matrix 1.2 ml 2D barcoded tubes (Thermo Fisher Scientific). Of the three 2D Matrix tubes only one was prefilled with the lysing matrix E (MP Biomedical), to which 100 µl of sample material was added. For the two other tubes, 800 µl of sample material was added and deposited at −20 °C in the MFD biobank. The tubes destined for downstream DNA extraction were added to a 96-well SBS rack containing 4 empty positions, 4 reaction blanks and 1 extraction positive control (https://github.com/SebastianDall/HT-Downscaled-Illumina-Metagenomes-Protocol). Linkage of the linear barcodes of the original sample container, the 2D Matrix tubes and location in the final SBS racks was ensured with the use of a Mirage Rack Reader (Ziath) and the software DataPaq (Ziath) forwarding the entries to an SQL server (MongoDB). Pseudolinks were generated for samples acquired as DNA extracts.
DNA extraction was performed using slightly modified protocol of the DNeasy 96 PowerSoil Pro QIAcube HT Kit (QIAGEN). In total, 500 µl CD1 was added to each 2D barcoded Matrix tube; the samples then underwent three 1,600 rpm bead-beating cycles performed at 2-min intervals using the FastPrep-96 (MP Biomedicals). Between the cycles, the samples were kept on ice for 2 min. After lysis, the samples were centrifuged at 3,486g for 10 min using an Eppendorf 5810 benchtop centrifuge (Eppendorf). Then, 300 µl supernatant was transferred by hand to a new S-block containing 300 µl CD2 and 100 µl nuclease-free water (NFW) to meet the requirement of 700 µl for the remaining part of the protocol. The samples were mixed by pipetting and centrifuged at 3,486g for 10 min; the sample transfer step was then performed using the QIAcube HT. All of the subsequent steps were performed according to the manufacturer’s protocol. DNA was quantified using the Qubit 1× HS assay (Invitrogen). Extraction metadata, including a denotation of methodology, can be found in Supplementary Data 6.
MFD ontology
For the MFD data, habitat classification was performed on site by experts in accordance with the relevant field guides when available for the habitats (that is, the natural habitats from macrobial ontologies). Habitat classification was therefore performed by checking the presence of plant indicator species, topographic and abiotic conditions. The MFDO was developed as a link between the classical plant-derived habitat ontologies and the Earth Microbiome Project Ontology (EMPO). The broadest MFDO classification level (Sample type), corresponds to the most specific EMPO level (EMPO level 4)3, while the detailed levels for natural samples correspond broadly to the Natura 2000 habitat ontology8. Finally, missing categories, such as urban, were adapted from the EUNIS ontology9 to provide a detailed description of non-natural habitats. The MFDO was designed, for the moment, to fit the Danish environment and it was refined with a panel of national experts. The full MFDO and its association to other habitat ontologies (that is, EMPO, Natura 2000 and EUNIS) can be found at GitHub (https://github.com/cmc-aau/mfd_metadata).
Metadata curation
The metadata collected with codeREADr were screened for completeness in the following fields (hereafter, minimal metadata): fieldsample_barcode (the unique sample identifier), project_id (unique identifier of the subproject), longitude and latitude (ISO 6709), sitename (common name of the sampling site), coords_reliable (indicating if the coordinates are reliable, not reliable or masked), sampling_date (sampling date ISO 8601) and five levels of the MFD habitat ontology (mfd_sampletype, mfd_areatype, mfd_hab1, mfd_hab2, mfd_hab3). If a sample presented an incorrect entry, (for example, wrong format, not meaningful for that column or unreadable), the error was corrected using R78 v.4.2.3, and if a correction was not possible, or the entry absent, the responsible person for the subproject was contacted. The process was iterated until improvements were not possible anymore. In brief, common corrections included case changing, date formatting (using lubridate79, v.1.9.2) and coordinate projection (project function from terra80, v.1.7.55). The reference grid mapping and masking of the coordinates were performed using the terra80 v.1.7.55 package. The European Environment Agency 1-km (ref. 81) and 10-km (ref. 82) reference grids of Denmark were projected from EPSG:3035 to EPSG:4326 (function project), whilst the coordinates from MFD samples were encoded into a spatial vector (function vect) and mapped onto the grids (function intersect) to identify their cells of origin. The cells associated with each sample (when the coordinates were present), were reported in the fields cell.1 km and cell.10 km of the metadata. The centroids of the cells were computed (function centroids) and, in the case of samples from subprojects P04_3 and P04_5, the centroids were provided as latitude and longitude, while the coords_reliable field for those samples was set to ‘Masked’. Coordinates from subproject P06_3 were provided already masked as generic locations in the commune of sampling. Concordance between manual annotation of the habitat and government-registered LU (land use) designation was inferred comparing the MFDO for each sample with the Basemap04 (ref. 10) aggregated LU map. A broad correspondence of terms between MFDO and LU terms was established and, to account for GPS and labelling inaccuracies, any match in a range of 20 m was considered in concordance. Samples that were in disagreement were screened manually on Google Maps and, if the disagreement was confirmed, the coords_reliable field was set to ‘No’. For Fig. 1, the base map was from EuroGeographics. This dataset includes Intellectual Property from European National Mapping and Cadastral Authorities and is licensed on behalf of these by EuroGeographics. The original dataset is available for free online (https://www.mapsforeurope.org). Terms of the licence are available at https://www.mapsforeurope.org/licence. All attribution statements can be found online (www.mapsforeurope.org/attributions). For Extended Data Fig. 1, the base of the map is from Eurostat (Geodata, GISCO, Eurostat; https://ec.europa.eu/eurostat/web/gisco/geodata).
Short-read metagenomic library preparation, sequencing and processing
Metagenomic libraries were prepared with a 1:10 reagent volume reduction of the standard Illumina DNA prep protocol (Illumina) as described previously77. Using the I.DOT One (DISPENDIX), 3 µl of up to 20 ng template DNA was prepared before addition of 2 µl BLT/TB1 and subsequent incubation in a thermocycler at 55 °C for 10 min. Tagmentation was stopped by addition 1 µl TSB using the I.DOT One, and incubation in the thermocycler at 37 °C for 15 min. The tagmented DNA was washed twice with 10 µl TWB. The I.DOT One was used to add the PCR master mix, prepared by mixing 2 µl EPM and 2 µl NFW per reaction. The epMotion 96 (Eppendorf) was used to add 1 µl IDT Illumina UD index (Illumina) to each reaction well. The input of genomic DNA was used to determine the applied cycles of the BLT-PCR program: 7 (4.9–20 ng), 8 (2.5–4.9 ng), 10 (0.9–2.5 ng) or 14 (< 0.9 ng). Size-selection was performed on the libraries by addition of 17 µl NFW, before 18 µl of the reaction volume was transferred to a new PCR-plate together with a mixture of 16:18 µl SPB:NFW. After incubation, 50 µl of the supernatant was transferred to a new PCR-plate with 6 µl undiluted SPB. After incubation, the beads were washed twice with 45 µl 80% ethanol and eluted in 20 µl NFW. SPB are 1:1 interchangeable with CleanNGS SPRI beads (CleanNA).
The individual libraries were quantified using a 1:10 diluted upper standard. The pooled libraries were concentrated using 2× volume of SPRI ProNex Chemistry (Promega) beads. Final sequencing libraries were produced by an equimolar combination of the pooled libraries. Quality control was performed using the Qubit 1× HS assay (Invitrogen) and DS1000 or DS1000 HS ScreenTape (Agilent Technologies). Library metadata are provided in Supplementary Data 6.
Metagenomic libraries were sequenced on the Illumina NovaSeq 6000 platform to a median depth of 5 Gb. If a library yielded insufficient data, the library was either repooled or reprepared for a second round of sequencing. The Illumina data were demultiplexed using bcl2fastq2 v2.20.0 (Illumina). The raw reads were trimmed for barcodes, quality filtered and deduplicated with fastp83 v.0.23.2 with the following options: --detect_adapter_for_pe --correction --cut_right --cut_right_window_size 4 --cut_right_mean_quality 20 --average_qual 30 --length_required 100 --dedup --dup_calc_accuracy 6. Commands were parallelized using GNU-parallel84 v.20220722 and outputs compressed using pigz85 v.2.4. Sequencing metadata are provided in Supplementary Data 6.
Nearly full-length bacterial 16S rRNA gene amplicon library preparation, sequencing and processing
A representative set of 426 samples were selected for 16S rRNA amplicon sequencing. These samples comprise 130 samples from the BIOWIDE project (P01_1)31, as well as 295 samples manually selected to reflect the sample diversity in the full dataset while attempting to maintain the geographical coverage. From samples from subterranean sediments (P12_1), the input DNA was pooled based on the sediment core the samples were derived from. From 14 of the samples, we failed to generate data of sufficient quality leading to a dataset of 412 samples. Here PCR was used to amplify the region V1–V8 of the 16S gene using UMI-tagged target primers enabling downstream chimera filtering and error-correction similar to the method described previously11. All of the samples were tagged by the UMI-tailed target primers lu_16S_8F and lu_16S_1391R in a PCR reaction (Supplementary Data 7). The reaction contained 10–20 ng DNA input, 1× SuperFi buffer, 0.2 mM dNTPs, 500 nM of each primer and 2 U of Platinum SuperFi DNA Polymerase (Invitrogen) in a total volume of 50 µl. The PCR program consisted of initial denaturation at 95 °C for 2 min followed by 2 cycles of denaturation (95 °C for 30 s), annealing (55 °C for 1 min) and extension (72 °C for 5 min). The PCR products were then purified with CleanNGS SPRI beads (CleanNA) at a ratio of 0.7× beads per sample. After 5 min of incubation, the beads were washed twice in 80% ethanol and eluted in 18 µl NFW for 5 min. The tagged molecules were then amplified in a second 25 cycle PCR reaction using barcoded primers targeting the UMI-adapter sequence. The PCR reaction contained 15 µl of the purified eluate, 1× SuperFi buffer, 0.2 mM dNTPs, 500 nM of forward and reverse primer and 2 U of Platinum SuperFi DNA Polymerase (Invitrogen) in a total volume of 50 µl. The PCR-program consisted of initial denaturation at 95 °C for 2 min followed by 25 cycles of denaturation (95 °C for 15 s), annealing (60 °C for 30 s) and extension (72 °C for 3 min), followed by a final extension at 72 °C for 5 min. The PCR products were purified with CleanNGS SPRI beads (CleanNA) as described above and eluted in 20 µl NFW. Poorly performing samples underwent a third PCR reaction with 5–10 cycles using up to 50 ng amplicon DNA as input and otherwise identical to the previous PCR reaction.
The barcoded amplicons were multiplexed in pools of 5–6 samples containing a total of 300 ng. The pools were used as input for library preparation for DNA sequencing using the ‘Amplicons by Ligation (SQK-LSK110)’ protocol (version: ACDE_9110_v110_revG_10Nov2020) and loaded onto a MinION R.9.4.1 flow cell (FLO-MIN106D). The flow cells were sequenced for up to 72 h on a GridION platform (Oxford Nanopore Technologies) using the MinKNOW software v.21.05.8 and basecalled using the super-accurate model (r941_min_sup_g507) with Guppy v.5.0.11. Downstream consensus sequences were generated using the longread_umi pipeline v.0.3.2 described previously11 with slight modifications to ensure compatibility with the updated medaka model (r941_min_sup_g507) and the custom barcode sequences. The quality of the consensus sequences was evaluated based on a ZymoBIOMICS Microbial Community DNA Standard (Zymo Research, D6306) included together with the samples. With UMI sequence coverage of ≥7× corresponding to Q30+ and ≥14× to Q40+. The exact command used to generate the consensus sequences was: longread_umi nanopore_pipeline -d input.fq -v 10 -o analysis -s 140 -e 140 -m 1000 -M 2000 -f GGAATCACATCCAAGACTGGCTAG -F AGRGTTYGATYMTGGCTCAG -r AATGATACGGCGACCACCGAGATC -R GACGGGCGGTGWGTRCA -c 3 -p 2 -q r941_min_sup_g507 -t 20 -T 2 -U “3;2;6;0.3”.
Bacterial and eukaryotic rRNA gene operon library preparation, sequencing and processing
A representative set of 450 samples was selected for both bacterial and eukaryotic rRNA operon sequencing. These samples are the same as those selected for 16S rRNA UMI amplicon sequencing. However, all extracted DNA had been used up for 8 of the samples, resulting in an overlap of only 404 samples, which is why we included 46 other samples. For bacterial rRNA sequencing, PCR was used to amplify around 4,500 bp targeting the 16S and 23S gene using the primers MFD_16S_8F and MFD_23S_2490R (Supplementary Data 7). For eukaryotic rRNA, operon sequencing PCR was used to amplify around 4,500 bp targeting the 18S and 28S gene using the primers MFD_18S_3NDF and MFD_28S_21R (Supplementary Data 7). The PCR reactions contained 10–20 ng DNA input, 1× SuperFi buffer, 0.2 mM dNTPs, 500 nM of each primer and 2 U of Platinum SuperFi DNA Polymerase (Invitrogen) in a total volume of 50 µl. With the addition of 1× SuperFi GC enhancer when amplifying the eukaryotic rRNA operons. The PCR-program consisted of initial denaturation at 98 °C for 1 min followed by 25 cycles of denaturation (98 °C for 15 s), annealing (55 °C for 15 s) and extension (72 °C for 3 min), followed by a final extension at 72 °C for 5 min. The PCR products were then purified with CleanNGS SPRI beads (CleanNA) at a ratio of 0.7× beads per sample. After 5 min of incubation, the beads were washed twice in 80% ethanol and eluted in 20 µl NFW for 5 min. The amplicon DNA was barcoded in a second 8–10 cycle PCR reaction using barcoded primers targeting the introduced adapter sequence. The PCR reaction contained 20 ng of the purified amplicon DNA, 1× SuperFi buffer, 0.2 mM dNTPs, 500 nM of forward and reverse primer and 2 U of Platinum SuperFi DNA polymerase (Invitrogen) in a total volume of 50 µl. With the addition of 1× SuperFi GC enhancer when amplifying the eukaryotic rRNA operons. The PCR program consisted of initial denaturation at 98 °C for 1 min followed by 8–10 cycles of denaturation (98 °C for 15 s), annealing (60 °C for 15 s) and extension (72 °C for 3 min), followed by a final extension at 72 °C for 5 min. The PCR products were purified with CleanNGS SPRI beads (CleanNA) as described above and eluted in 18 µl NFW.
The barcoded amplicons were multiplexed in pools of 92 samples containing a total of 1–2 µg of DNA. The pools were size-selected using SPRI ProNex Chemistry (Promega) with a ratio of 1.2× beads per sample according to the manufacturer’s protocol and eluted in 125 µl. The size-selected and purified pools were shipped for PacBio CCS sequencing on the Sequel II platform (Pacific Biosciences) using the binding kit 3.2. The CCS sequences were further processed using Lima v.2.6.0 (Pacific Biosciences) to filter and demultiplex the data. This was done using the hifi-preset ASYMMETRIC and the following settings --min-score 70, --min-end-score 40, --min-ref-span 0.75, --different, --min-scoring-regions 2. Furthermore, remaining ligation products were removed by identifying partially remaining adapter sequences after filtering. Subsequently all reads were oriented while removing primer sequences and filtering reads below 3.5 kb or above 6.5 kb using cutadapt86 v.3.4. 16S rRNA genes corresponding to the V1–V8 region were extracted from the rRNA operons using a custom script (trim_RNA_operons.sh) that carries out several steps: first, the rRNA operons were truncated to 1,450 bp using the usearch87 v.11 command fastx_truncate -trunclen 1450. The trimmed sequences were then trimmed based on the 1391R88 (Supplementary Data 7) primer using cutadapt86 v.3.4. The sequences for which the primer could not be found were aligned to the global SILVA v.138.1 SSURef NR99 (ref. 19) alignment using SINA89 v.1.6.0, the aligned sequences were trimmed according to the position of the primer binding sites in the alignment. Truncated sequences were removed using a custom script (remove_incomplete_seqs_from_sina_aln.py) that considered sequences that start or end with three or more gaps as truncated. Finally, gaps were removed using the custom script (Remove_gaps_in_fasta.py), whereafter the primer- and alignment-trimmed sequences were combined.
Before diversity analysis, 18S rRNA genes, corresponding to the position between the 3NDf90 and 1510R91 primer-binding sites (Supplementary Data 7), were extracted using a custom script (trim_euk_RNA_operons_3ndf-1510R.sh). This script was identical to the script used for processing bacterial rRNA operons except that sequences were trimmed based on the 1391R88 primer (Supplementary Data 7) with cutadapt86 v.3.4 and the alignment trimmed based on the corresponding position in the SILVA19 global alignment using SINA89 v.1.6.0. The resulting reads were dereplicated using usearch87 v.11 command usearch -fastx_uniques -sizeout and then resolved into ASVs using usearch -unoise3 -minsize 2. The phylogenetic diversity of the 18S rRNA genes was determined by clustering the ASVs into OTUs at 99% identity with usearch -cluster_smallmem -maxrejects 512 -sortedby other, followed by mapping against the PR2 (ref. 16) database to determine the percentage identity with the closest hit in the database using usearch -usearch_global -maxrejects 0 -maxaccepts 0 -top_hit_only -id 0 -strand plus. An OTU-table was created by mapping the trimmed raw reads against the 99% OTUs using usearch -otutab -otus. Taxonomy was assigned to the OTUs using the UTAX version of the PR2 database16 v.5.0.0, the SINTAX classifier through usearch63,87 v.11. For 18S diversity analyses, the taxonomy was inferred with the DADA2 (ref. 92) v.1.26.0 function assignTaxonomy. OTUs not classified as Eukaryota were discarded before the downstream analyses.
Estimation of the Danish terrestrial diversity
The nearly full-length 16S rRNA UMI gene sequences were mapped against the nearly full-length OTUs clustered at 98.7% sequence similarity to yield a dataset (OTU table) with 412 habitat-representative samples and 107,826 (106,760 after filtering) species-representative OTUs using usearch87 v.11 with downstream analysis done in R78 v.4.4.1 using tidyverse93 v.2.0.0. Similarly, the full-length 18S rRNA gene sequences were mapped against the nearly full-length OTUs clustered at 99% sequence similarity, yielding a dataset of 450 habitat-representative samples and 12,515 (12,469 after filtering) species-representative OTUs.
We made habitat-specific rarefaction curves across samples with more than 4,000 observations from habitats with more than 9 sample representatives (5.9 million observations), as well as a pan-habitat rarefaction curve (6.0 million observations) from the nearly full-length bacterial rRNA gene UMI dataset. For this analysis, we combined the samples from temperate heath and scrub (n = 12) and sclerophyllous scrub (n = 7). Likewise, we made habitat-specific rarefaction curves across samples with more than 6,000 observations from habitats with more than 9 sample representatives (12.0 million observations), as well as a pan-habitat rarefaction curve (12.9 million observations) from the eukaryotic nearly full-length 18S rRNA gene dataset. For this analysis we combined the samples from temperate heath and scrub (n = 13) and sclerophyllous scrub (n = 7). Rarefaction was performed using vegan94 v.2.6-6-1 (function rarecurve) using a step size of 10,000.
Sample based coverage and Hill diversity indices were calculated after transformation to presence and absence data by using rarefaction and extrapolation with Hill numbers of order q as implemented in the iNEXT95 v.3.0.1 package (function iNEXT). Hill richness (total number of species), Hill–Shannon (number of common species) and Hill–Simpson (number of dominant species) were estimated using order q = 0, q = 1 and q = 2, respectively. For the total dataset, the end point of extrapolation was set to twice the size of the dataset (nBac = 824, nEuk = 900); and, for habitat-specific estimates, the end point was fixed at 100 samples. For the habitat-specific data, we investigated normality (function shapiro_test) of the community coverage and log-transformed sampling effort and, based on the results, measured the linear correlation with the Pearson correlation coefficient (function cor_test) using the package rstatix96 v.0.7.2.
Establishment of the MFG 16S rRNA gene reference database
The MFG 16S rRNA reference database was assembled from high-quality bacterial and archaeal 16S rRNA genes obtained from several sources: nearly full-length 16S rRNA gene and rRNA operon amplicons created in this study, SILVA v138.1 SSURef NR99 (ref. 19), EMP500 (ref. 3), AGP70 (ref. 11), MiDAS 4 (ref. 97) and MiDAS 5 (ref. 20), and ref. 21. All bacterial sequences were trimmed between the 8F98 and 1391R99 primer-binding sites (Supplementary Data 7), and archaeal sequences between the 20F100 and the SSU1000ArR101 primer-binding sites (Supplementary Data 7).
High-quality bacterial and archaeal sequences were obtained from the SILVA v138.1 SSURef NR99 (ref. 19) ARB-database by exporting them separately in the fastawide format after terminal trimming between positions 1,044 and 41,788 (Bacteria) and positions 1,041 and 32,818 (Archaea) in the global SILVA19 alignment, corresponding to the primer binding sites (Supplementary Data 7). A custom script (Extract_full-length_16S_rRNA_genes_from_SILVA_alignments.sh) was used to remove truncated sequences based on the presence of terminal gaps in the exported FASTA-alignments. Finally, sequences that contained N’s were removed and U’s were replaced with T’s using two custom scripts (Remove_seqs_with_Ns.py and replace_U_with_T.py).
Owing to the large number of trimmed 16S rRNA gene reads, ASVs were resolved for each dataset individually. Sequences were dereplicated using the usearch87 v.11 command usearch -fastx_uniques -sizeout and then resolved into ASVs using usearch -unoise3 -minsize 2. ASVs from all individual datasets were combined with the 16S sequences from SILVA v138.1 SSURef NR99 (ref. 19) and dereplicated using usearch -fastx_uniques. The ASVs were sorted based on their abundance across the MFD dataset by mapping the trimmed sequences against the ASVs with usearch -search_exact -dbmask none -strand plus -matched. The matched sequences were dereplicated using usearch -fastx_unique -sizeout, and sequences which did not originate from the MFD samples were appended. The complete database was processed using AutoTax18 v.1.7.6 to create the MFG 16S reference database. As the MFG database was found to contain sequences representing chloroplast and mitochondrial 16S rRNA genes, as well as pseudogenes, we performed additional filtering to create the final MFG database. To inform this filtering, we undertook extensive manual evaluation of the phylogenetic tree and sequence alignments in ARB102 v.7.0 to establish a reproducible method for removing non-16S rRNA gene sequences. These steps included removing (1) all sequences that shared less than 70% identity with their closest match in the SILVA v138.1 SSURef NR99 database19, as these most likely represent pseudogenes and would lead to inflated phylum level diversity; (2) ASVs with de novo placeholder names and best hits in SILVA v138.1 SSURef NR9919 against Mitochondria or Chloroplast; (3) ASVs with de novo phyla placeholder names and top hits against Rickettsiales and less than 75% identify, as these also probably represent mitochondrial sequences that are not represented in the SILVA database; (4) ASVs representing de novo phyla covered by only a single ASV; (5) ASVs with a better hit in the MIDORI2 GB257 mitochondrial database103 than in SILVA19 v138.1 SSURef NR99, which, at the same time, share >75% identity (> 1,000 bp alignment length) to the MIDORI2 GB257 (ref. 103) hit. Finally, ASVs that were assigned to de novo phyla but shared >70% identity with a Patescibacteria hit were assigned to Patescibacteria. The species-level clustered (98.7% identity) version of the MFG 16S reference database was created by subsetting the MFG 16S reference database to those representing the 98.7% clustering centroids created during the AutoTax18 processing based on their ASV numbers.
Database evaluation
To evaluate the coverage of the MFG database, we classified 16S gene fragment reads (13.1 million) extracted from representative metagenomes based on the 10 km EU reference grid of Denmark (see the ‘Spatial thinning’ section) and 16S rRNA gene V4 OTUs clustered at 99% identity (2.26 million) from the GPC project23 using our database, as well as SILVA 138.1 SSURef NR99 (ref. 19), GreenGenes2 (ref. 22) and the complete 16S rRNA database from GTDB R22034 using the SINTAX classifier63, after which, the percentage of reads classified at different taxonomic ranks was determined. The analysis was conducted using R78 v.4.3.2 using tidyverse93 v.2.0.0 and vegan94 v.2.6-6.1.
Short-read 16S rRNA gene classification
Hidden Markov models (HMM) were made from Rfam104 v.14.7 seed alignments for Archaea (RF01959), Bacteria (RF00177) and Eukarya (RF01960) using hmmbuild (HMMER105 v.3.3.2). Metagenomic reads from rRNA gene fragments were extracted from the quality-filtered metagenomic reads using the constructed models with nhmmer106 (HMMER v.3.3.2) with the settings --incE 1e-05 -E 1e-05 --noali. In the case of multiple hits for the same metagenomic read, the best domain hit was selected based on the bit-score. The 16S reads were filtered for hits within the region between position 8F98 and 1391R99 primer binding sites for Bacteria (Supplementary Data 7) and the 20F100 and SSU1000ArR101 primer binding sites for Archaea (Supplementary Data 7). The 16S reads were taxonomically annotated using the SINTAX classifier63 and the MFG 16S reference database with the confidence cutoff set to 0.8. The output was aggregated to the individual taxonomic levels using R78 v.4.4.1 and the tidyverse93 v.2.0.0 package. Reads not classified at the given taxonomic level were assigned to ‘unclassified’.
Spatial thinning
To investigate the effect of spatial autocorrelation on community composition, we performed a distance decay analysis across the levels of the MFD ontology. The spatial autocorrelation was modelled using a logarithmic decay model of Hellinger-transformed Bray–Curtis similarity (1 − Bray–Curtis dissimilarity) as a function of spatial distance. Spatial distances were calculated from longitude and latitude with the codep107 v.1.2-3 package (function geodesics) using the Haversine formula. A pseudocount of 0.1 m was added between samples with a spatial distance of zero. Community similarity was calculated using the metagenomic 16S rRNA gene fragments aggregated at the genus level from samples with more than 1,000 observations. This led to a dataset with 10,001 samples and 38.4 million observations, which was subjected to random subsampling without replacement to a depth of 1,002 observations using ampvis2 (ref. 108) v.2.8.9 (functions amp_load and amp_subset_samples). After transformation, relative abundances were Hellinger-transformed using vegan94 v.2.6-6.1 (function decostand) before calculation of Bray–Curtis dissimilarity using the package parallelDist109 v.0.2.6 (function parDist) and summarized using hexbin110 v.1.28.3. The modelling was restricted to MFDO1 habitats showing spatial separation of samples, excluded Bornholm, and was limited to spatial distances ≤300 km. These filtering criteria lead to the inclusion of 9,121 samples. We made individual models for comparisons within and between the same habitat categories across all five levels of the habitat ontology. We found that at 10 km and on the MFDO1 level and above, the spatial effect becomes negligible. To address the effect of spatial autocorrelation on community composition we made a spatially thinned dataset using the 10-km reference grid of Denmark82. For each MFDO1 habitat, we chose sample representatives by selecting the sample with the lowest mean Bray–Curtis dissimilarity to the other samples of that MFDO1 habitat in the same cell. This led to a spatially thinned subset of 2,348 samples.
Effect of drying on diversity metrics and community composition
We performed 16S rRNA gene amplicon sequencing of the V4 region on replicate samples originating from two different lots at an agricultural experimental station. The samples were processed as triplicates which were either subsampled the day of collection and immediately frozen and stored in the freezer for +6 months and then subsampled, or dried at either room temperature (25 °C), 40 °C, 60 °C or 80 °C overnight and then stored for +6 months. The dried samples were rewetted with PBS before subsampling and DNA extracted using the DNeasy 96 PowerSoil Pro QIAcube HT Kit (QIAGEN) as described above. We investigated normality (function shapiro_test) and homoscedasticity (function levene_test) of the amount of extracted DNA using the package rstatix96 v.0.7.2. Based on the results obtained, we chose a nonparametric approach to test the significance of the differences. We used a Kruskal–Wallis rank sum test (function kruskal_test) combined with two-sided post hoc pairwise Mann–Whitney U-test comparisons (function wilcoxon_test) from rstatix96 v.0.7.2. The Bonferroni procedure was applied to adjust for multiple testing.
DNA was diluted to 5 ng μl−1 using NFW and standard amplicon libraries were prepared as described previously77. In brief, the amplicon libraries were prepared as one 50 μl reaction, which was subsequently split into two 25 μl reactions. Then, 25 cycles of PCR were performed on the duplicate samples, which were subsequently pooled and cleaned using 0.8× CleanNGS SPRI beads (CleanNA) and washed twice with 80% ethanol and eluted in NFW. Another 8 cycles of library PCR were performed on up to 10 ng of amplicon template and cleaned with 0.8× CleanNGS SPRI beads (CleanNA). The final libraries were quantified and pooled equimolarly to produce the final sequencing libraries. Each 25 μl amplicon PCR reaction consisted of 4 μl sample/NFW (target: 20 ng DNA), 1 µl UV-treated NFW, 25 μl PCRBIO 2× Ultra Mix and 20 μl abV4-C tailed amplicon primer mix (1 μM, 400 nM final concentration). The subsequent 25 μl library PCR was prepared with the cleaned PCR template in 2 μl sample/NFW (target: 10 ng DNA), 0.5 µl NFW, 12.5 μl PCRBIO 2× Ultra Mix (PCR Biosystems) and 10 μl adapter indexes (4 μM). After clean-up, libraries were pooled equimolarly. A detailed protocol can be found at GitHub (https://github.com/SebastianDall/HT-downscaled-amplicon-library-protocol).
The final library was sequenced on the Illumina MiSeq platform. ASV abundance tables were generated by running AmpProc v.5.1 (https://github.com/eyashiro/AmpProc) using the following choices: standard workflow, generate both otu and zotu tables, process only single-end reads, no primer region removal, amplicon region V4 and no taxonomic assignment. The 16S reads were taxonomically annotated using the SINTAX classifier63 and the MFG 16S reference database with the confidence cutoff set to 0.8.
We filtered the data to samples with more than 10,000 observations, which led to a dataset of 35 samples out of the original 36 and a total of 1.65 million observations. We performed random subsampling without replacement to a depth of 36,975 observations using ampvis2 (ref. 108) v.2.8.9 (functions amp_load and amp_subset_samples).
We used ampvis2 (ref. 108) v.2.8.9 to investigate community composition (function amp_heatmap) and to perform exploratory ordination analysis (function amp_ordinate) using Hellinger-transformed Bray–Curtis dissimilarities of ASVs with more than 0.01% relative abundance in any sample. We used ampvis2 (ref. 108) v.2.8.9 to calculate observed ASV richness (function amp_alphadiv) and Bray–Curtis dissimilarity (function vegdist) after Hellinger-transformation (function decostand) of relative abundances using vegan94 v2.6-6.1. We investigated normality (function shapiro_test) and homoscedasticity (function levene_test) of both observed ASV richness and Bray–Curtis similarity (1 − Bray–Curtis dissimilarity) using the package rstatix96 v.0.7.2. On the basis of the results obtained, we chose a nonparametric approach to test the significance of the differences. We used a Kruskal–Wallis rank-sum test (function kruskal_test) combined with two-sided post hoc pairwise Mann–Whitney U-test comparisons (function wilcoxon_test) from rstatix96 v.0.7.2. The Bonferroni procedure was applied to adjust for multiple testing. We performed a PERMANOVA to access the effect of treatment on the community composition (as measured as Hellinger-transformed Bray–Curtis dissimilarity) and partitioned the variance by contrasts using vegan94 v.2.6-6.1 (function adonis2). In both cases 9,999 permutations were used.
Evaluate the variable treatment effect in agricultural soils from MFD
We selected samples in the metagenomic 16S rRNA gene fragment dataset from either the pool of dried samples (n = 2.617) or frozen samples (n = 385) but excluded the frozen samples from lowland soils. To ensure spatial separation, we selected the representative samples from the spatially thinned subset while considering the different type of crops. This procedure led to the selection of 30 dried and 30 frozen samples. The sampling points were visualized on a map of Denmark using the ggplot111 extension ggspatial112 v.1.1.9 and the base map from rnaturalearth113 v.1.0.1. We performed a principal coordinate analysis (PCoA) of Hellinger-transformed Bray–Curtis dissimilarities. Principal coordinate decomposition performed with the ape114 v.5.8 package (function pcoa). We evaluated dispersion of the habitats using vegan94 v.2.6-6-1 (function betadisper). The significance of the overall dissimilarity between habitats was evaluated using ANOSIM (function anosim) using vegan94 v.2.6-6-1 with 9,999 permutations. We performed a PERMANOVA to access the effect of treatment on the community composition using vegan94 v.2.6-6.1 (function adonis2) with 9,999 permutations.
Diversity metrics
Based on the sampling methodology and DNA extraction summaries, we limited the analysis of diversity metrics to surface sediment and topsoil from environments with a minimum of nine sample representatives. We filtered the data to samples with more than 4,000 and 6,000 reads in the nearly full-length 16S rRNA gene UMI dataset and the nearly full-length 18S rRNA gene dataset respectively. For the diversity calculations, we combined the samples from ‘temperate heath and scrub’ and ‘sclerophyllous scrub’. We performed random subsampling without replacement to a level of 4,008 and 6,235 for the 16S and 18S rRNA gene data, respectively using vegan94 v.2.6-6-1 (function rrarefy). After this procedure, the nearly full-length 16S rRNA UMI dataset comprised 309 habitat-representative samples, 76,052 species-representative OTUs and 1.2 million observations, while the numbers for the nearly full-length 18S were 363 samples, 10,356 species representative OTUs and 2.2 million observations. Finally, the nearly full-length 18S rRNA gene data were transformed to presence and absence.
For the prokaryotic communities, the beta diversity was evaluated from the Bray–Curtis dissimilarity matrix (see the ‘Spatial thinning’ section) calculated from the metagenome-derived 16S rRNA gene fragment dataset. After subsetting to the spatially thinned set of samples and filtering to the environments used in analysis of alpha and gamma diversity, the matrix encompassed dissimilarities between 1,954 samples. For the eukaryotic communities, we calculated Jaccard dissimilarity on the nearly full-length 18S rRNA using the package parallelDist109 v.0.2.6 (function parDist) due to sparsity of 18S rRNA gene reads in the metagenomic data.
Alpha diversity was calculated as observed OTU richness using ampvis2 (ref. 108) v2.8.9 (function amp_alphadiv). We investigated normality (function shapiro_test) and homoscedasticity (function levene_test) of the groups using the package rstatix96 v.0.7.2. Owing to multiple cases of non-normality, a nonparametric approach was applied. Significance of differences in the observed richness was statistically tested using a Kruskal–Wallis rank-sum test (function kruskal_test) and two-sided post hoc pairwise Mann–Whitney U-test comparisons (function wilcoxon_test) from rstatix96 v.0.7.2. The Benjamini–Hochberg procedure was applied to adjust for multiple testing. A compact letter display was made using the function multcompLetters from package multcompView115 v.0.1-10 using a confidence threshold of 0.05. Gamma diversity was estimated after transforming the prokaryotic data to presence and absence, by using rarefaction and extrapolation with Hill numbers of order q as implemented in the iNEXT95 v.3.0.1 package (function iNEXT). The end point of extrapolation was fixed at 100 samples and gamma diversity reported as Hill–Shannon using order q = 1. Differences between groups were inferred from the 95% confidence intervals. Beta diversity was evaluated at both mean within-habitat and mean between-habitat level. Differences in beta diversity were visualized using the stats78 v.4.4.1 package (function hclust, method ward.D2) on the between-habitat Bray–Curtis dissimilarities, with the confidence of the habitat-splits calculated using 100 iterations with the bootstrap116 v.0.1 package (function bootstrap). The analysis was conducted using R78 v.4.4.0 using tidyverse93 v.2.0.0.
Exploratory PCoA
We explored differences in community composition between different habitats by performing PCoA on the Bray–Curtis dissimilarity matrix calculated from the metagenome-derived 16S rRNA gene fragment dataset (see the ‘Spatial thinning’ section) and from the Jaccard dissimilarity matrix of nearly full-length 18S rRNA gene dataset of presence and absences. For the prokaryotic metagenomic-derived data, we removed samples from subterranean environments, samples from drinking water treatment plants and habitats with less than 20 sample representatives, the matrix encompassed Bray–Curtis dissimilarities between 9,643 samples. For the nearly full-length eukaryotic data, we used the same data as for calculations of alpha and gamma diversity resulting in a matrix of Jaccard dissimilarities between 363 samples. Principal coordinate decomposition was performed with the ape114 v.5.8 package (function pcoa). We evaluated dispersion of the habitats using vegan94 v.2.6-6-1 (function betadisper). The significance of the overall dissimilarity between habitats was evaluated using ANOSIM (function anosim) using vegan94 v.2.6-6-1 with 999 permutations. To evaluate how much of the variance could be explained by the MFDO1 habitat levels we performed a PERMANOVA (function adonis2) using vegan94 v.2.6-6-1 with 999 permutations. We expanded the analysis using a contrasts analysis also using adonis2 and 999 permutations.
Habitat classification
The habitat classification analysis was performed using R78 v.4.2.3 according to the workflow described in Supplementary Note 5. The microbial relative abundances at the genus level of the 16S gene fragments were summarized at higher taxonomic levels (family to phylum) by summing up the abundances of populations with the same taxonomy. Taxa observed with a relative abundance >0 in at least 25 samples were retained for subsequent analysis. The resulting five tables were screened for multicollinearity using the function vifcor (th = 0.7) from the package sdm117 v.1.1_18 wrapped in a block-wise script for efficiency. Considering the block-wise implementation, it is not guaranteed to find the optimal solution, but the shuffling at each iteration increases the chances of approaching it. The microbial data were filtered for a minimum (n = 25) of species observations, spatially thinned (by habitat class) with a minimum distance of 5 km using the function thin_by_dist from tidysdm118 v.0.9.5. The ontology was used to create five different target variables, one for each ontology level, but only classes with at least 50 observations and whose class names were not ending with ‘NA’ were retained for modelling. The modelling was carried out with the package tidymodels119 v.1.1.1, which allows the creation of model workflows. The functions cited in this section belong to tidymodels119 or its dependencies unless otherwise stated. The data were split (stratified by class) in training and test set (70/30 split) using the function initial_split and, in the training data (70% of the filtered set), the minority classes were upsampled using the SMOTE algorithm from the package themis120 v.1.0.3 with a ratio equal to 0.5. The predictor variables (microbial relative abundances) were centred on the mean, scaled to unit variance and used to build a random forest model (from the package ranger121 v.0.16.0). The models’ hyperparameters mtry, trees and min_n were tuned using a fivefold cross validation using the function vfold_cv with v = 5 and repeats = 5; leading to 25 fits from fit_resample and an equal number of evaluation points per hyperparameters’ combination. The best model was selected and evaluated on the test set (30% of filtered data). Moreover, the variable importance (impurity) was retrieved for the best model. Several steps in the workflow involve random choices (Supplementary Note 5), therefore, to smoothen the effects of randomness, the workflow was repeated 25 times starting from the spatial thinning. Global metrics such as F1 (micro and macro), PR-AUC and Kappa were collected after the validation using the function collect_metrics. We collected a total of 25 best models, one for each combination between predictors (phylum, class, order, family, genus) and targets (sample type, area type, MFDO1, MFDO2, MFDO3). The analysis of the false negatives was performed by collecting, for each class in each fold and iteration the number of associated misclassified samples, using the function conf_mat from the package yardstick122 v.1.3.0. A null distribution of false negatives was computed by multiplying the previous number by the fraction of samples in each class (except for the class in exam). A two-tailed paired t-test was performed using the function t_test from the package rstatix96 v.0.7.2.
Core analysis
Abundant and prevalent genera were identified from the spatially thinned dataset across each level of the MFD ontology using a prevalence and relative abundance filter of 50% and 0.1% respectively. Investigation of shared genera was performed using UpSetR123 v.1.4.0 and ComplexUpset124 v.1.3.3. We screened the core genera for candidates related to habitat disturbance and visualized their abundance and prevalence across the different MFDO1-level habitats. We mapped all 16S rRNA genes from metagenomic bins (see below) against the MFG 16S reference database (98.7% identity) using usearch87 v.11 usearch_global, with the flags -top_hits_only and -strand both. We then identified genera associated with nitrogen cycling and investigated their abundance and prevalence across the MFDO1 habitats.
Metagenomic assembly and binning
Trimmed shallow metagenomic reads were assembled using MegaHit125 v.1.2.9 with the following options: --k-list 27,43,71,99,127 --min-contig-len 1000. Metagenomes smaller than 1 Mb were omitted from further processing. In total, 10,042 assemblies were used for genome recovery.
To maximize the recovery of low coverage MAGs, the SingleM40 v.1.0.0beta7 ‘pipe’ subcommand with the default GTDB R214 metapackage126 was first run on the metagenomes to generate archive OTU tables of single-copy marker gene sequences (combined with SingleM summarize across all samples). Bin Chicken127 v.0.9.6 was then run on those tables using the command ibis coassemble –max-coassembly-samples 1 –max-recovery-samples 10 –singlem-metapackage S3.2.1.GTDB_r214.metapackage_20231006.smpkg.zb, to match the single-copy protein sequences across samples and choose the 10 most similar samples for each sample for multisample binning. The reads for the selected samples were mapped to the assemblies using Minimap2 (ref. 128) v.2.24 with the -ax sr option and SAMtools129 v.1.16.1 with samtools view -Sb -F 2308 - | samtools sort options. The jgi_summarize_bam_contig_depths command of MetaBAT2 (ref. 130) v.2.12.1 was used on the mapping files to acquire contig coverage values, which were used as input for binning through MetaBAT2 (ref. 130) with the following options: -m 1500 -s 100000.
The recovered MAGs were quality-assessed using CheckM2 (ref. 131) v1.0.2 to acquire MAG completeness, contamination values and were taxonomically classified using GTDB-Tk132 v.2.4.0. Bakta133 v.1.8.1 with Bakta database v.5.0 (type full) was used to annotate the MAGs and acquire bacterial rRNA and tRNA counts, while for archaeal MAGs, tRNAscan-SE134 v.2.0.9 and barrnap135 v.0.9 with the corresponding archaeal databases were used to acquire tRNA and rRNA counts. CheckM2 (ref. 131) completeness and contamination values, together with the observed rRNA and tRNA counts, were used to classify the MAGs according to MIMAG136 guidelines, and only medium- or high-quality MAGs were kept for further analysis. The MAGs were dereplicated using dRep137 v.2.6.2 with the -sa 0.95 -nc 0.4 settings. MAG coverage values were calculated using CoverM138 v.0.6.1, with the -m mean setting.
Representation of the metagenomic microbial community by these short-read-assembled MAGs in each sample was assessed using SingleM40 v.0.18.0 (default GTDB R220 reference metapackage139 v.4.3.0) through the appraise subcommand that compares the OTUs of 59 single-marker copy genes found in the metagenomic reads and MAGs. In brief, the SingleM pipe subcommand was first applied to individual trimmed short-read metagenomes (with a minimum of 100 bp) and individual short-read assembled MAGs to identify OTU sequences of these 59 marker genes in both types of datasets. For each type of dataset (that is, short-read data and MAGs), OTU tables were then combined using SingleM summarize and passed through the SingleM appraise subcommand (under the default cut-off for species-level estimates) to compare the OTU tables of both data types for determining the bacterial and archaeal community recovered by the short-read assembled MAGs.
For ‘Candidatus Nitrososappho danica’ and ‘Candidatus Nitronatura plena’, we conducted a targeted reassembly to retrieve the highest quality possible genome representatives for the type material. Long-read assemblies of MFD06229 (containing ‘Candidatus N. natura’) and MFD09848 (containing ‘Candidatus Nitrososappho danica’) were taxonomically classified with mmseqs2 (ref. 140) v.14.7e284 using the uniref100 database141 (12 October 2024 download date). For reassembling MFD06229.bin.1.58 (NCBI: GCA_974707355.1), reads mapping to contigs classified as the Nitrospirota phylum were extracted using the Samtools129 v.1.20 “view -q 20 -m 1000” command and assembled with myloasm142 v.0.1.0. The acquired circular contig was extracted as a separate genome bin. For MFD09848.bin.1.115 (NCBI: GCA_974504955.1), reads mapping to contigs of the Nitrososphaerota phylum were extracted with the Samtools view -q 20 -m 1000 command and assembled using Flye143 (v.2.9.3) with the following settings: --nano-hq, --meta, --extra-params min_read_cov_cutoff = 12. A MAG was then recovered through manual binning and curation by comparing the reassembled contigs to the original MFD09848.bin.1.115 genome.
Novelty and prokaryotic fraction estimation
To assess sample novelty in each shallow metagenome, microbial profiles were estimated by SingleM40 v.0.18.0 (default GTDB R220 reference metapackage v.4.3.0) using the SingleM pipe subcommand. Essentially, OTUs of 59 single-copy marker genes were first identified from the shallow metagenomes and compared against those found in the reference genome database to assign taxonomic classifications for the identified OTUs. The overall taxonomic profile for each metagenome was then condensed from the OTU tables of all the different markers. Sample novelty was expressed in terms of the proportion of microbial community characterized by the current reference genome database that encompassed most recent advances in genome mining. This was achieved by taking the ratio between the sum of the total coverage at the species level and the sum of total coverage at all taxonomic levels based on the condensed taxonomic profiles (hereafter, known species fraction). To compare novelty between metagenomes from MFD and NCBI public metagenomes, published SingleM40 profiling results for NCBI datasets were used for comparison with the MFD datasets. NCBI datasets were grouped into NCBI_Water, NCBI_Soil, NCBI_Sediment and NCBI_Human based on the original metadata labelling in the original SingleM publication using the habitat mapping in Supplementary Table 3. NCBI_Human refers to the human gut metagenomes in NCBI, and they were used as a reference point for a well-studied system.
To investigate the improved species representation of the microbial community by the short-read assembled MAGs, the MAGs were added to the default GTDB R220 SingleM metapackage139 v.4.3.0. This was done using the SingleM supplement subcommand applying dereplication at 95% similarity and CheckM2 (ref. 131) quality filtering of 50% completeness and 10% contamination for all MAGs to be supplemented. In total, 4,507 dereplicated and novel species representatives from 19,253 MFD MAGs were added to the new metapackage. The microbial community was then reanalysed based on this new metapackage supplemented with the short-read constructed MAGs using the SingleM renew subcommand.
Genome quantification
We quantified the microorganisms across Denmark using the MFD MAG catalogue as reference and the full set of short-read metagenomes with sylph41 v.0.6.1. First, we indexed the reference and the samples using the sketch command and the default parameters. The command profile was used to profile the sample sketches against the genome sketches. We extended the microbial taxonomy from GTDB34 R220 to include the dereplication cluster identifier calculated by dRep137 v.2.6.2 (see the ‘Metagenome assembly and binning’ section). We incorporated the microbial taxonomy using the utility script sylph_to_taxprof.py and then extracted the quantification matrices with the utility script merge_sylph_taxprof.py. The taxonomic abundance was aggregated from individual genome to species clusters in R78 v.4.2.3 using the tidyverse93 v.2.0.0 package. For analysis of genome abundances of AOA species cluster TA-21 sp02254895 cluster 98_1, one sample was randomly selected from each 10 km reference cell within each MFDO3 level. Only MFDO3 levels represented in at least 10 different 10 km reference cells were included in the analysis, to exclude habitat types only being present in a confined area of Denmark. We investigated normality (function shapiro_test) and homoscedasticity (function levene_test) of the groups using the package rstatix96 v.0.7.2. Owing to non-normality, a nonparametric approach was applied for comparison of groups. Pairwise comparison was performed between each group (MFDO3) against all (base mean) using a one-sided Mann–Whitney U-test (function wilcoxon_test).
Gene-centric investigation of short-read metagenomes
A dereplicated set of the species representatives from the GTDB34 release 214 and the genomes from Earth’s microbiomes4 GEM database was created using fastANI144 v.1.32 removing all GEMOTU genomes with >96% average nucleotide identity over >50% aligned fragments to a GTDB34 species representative. The resulting 97,227 genomes (85,205 GTDB and 12,022 GEMOTU) were annotated using anvi’o145 v.8 with gene calling using Prodigal146 v.2.63.
The total protein complement of the 97,227 genomes was used as a query in a DIAMOND147 v.2.1.8 search against preliminary protein datasets of cytoplasmic NarG/NxrA, the two distinct versions of periplasmic NxrA/NarG (exemplified by Nitrospira and Nitrotoga, respectively), and AmoA/PmoA with a score cut-off of 100. True-positive hits were selected using an alignment score ratio approach as previously described148,149, resulting in 1,068 AmoA/PmoA sequences, and 10,254 cytoplasmic NxrA/NarG sequences, 665 periplasmic NxrA/NarG sequences of the Nitrospira clade, and 581 periplasmic NxrA/NarG sequences of the Nitrotoga clade. To search the short-read metagenome data for AmoA and NxrA/NarG functional genes, we created several new GraftM39 v.0.14 search packages: Archaeal AmoA, Bacterial AmoA, cytoplasmic NxrA/NarG and periplasmic NxrA/NarG. The AmoA/PmoA sequences were split into 823 bacterial and 245 archaeal sequences to create two specific GraftM packages. Moreover, the two periplasmic NxrA/NarG sequence databases were combined to make one GraftM package.
Multiple-sequence alignment of protein sequences was performed with MAFFT150 v.7.490. TrimAl151 v.1.4.1 was used to trim minimum 20% amino acid representation. IQ-TREE152 v.2.2.0.3 was used to generate phylogenetic trees of protein sequences, using the ultrafast bootstrap approximation option and 1,000 iterations. For NxrA/NarG and bacterial AmoA trees, the ModelFinder153 option was applied (best-fit models pNXR: LG + R6, cNXR: LG + F + R10, Bacterial amoA: LG + F + R8). The model applied for the archaeal AmoA tree was WAG + R. ARB102 v.7.0 was used to reroot and group trees, followed by visualization in iTOL154 v.6. Trees used for GraftM39 v.0.14 package generation are shown in Supplementary Fig. 10. Inclusion of protein sequences of NxrA and AmoA obtained through this study was filtered based on length of the protein product, with a cut-off of 200 amino acids for AmoA and 600 amino acids for NxrA. Gene phylogeny was ascertained using GraftM39 v.0.14 on the forward shallow metagenomic reads with search mode hmmsearch+diamond along with a conditional E-value threshold of 10−10. The samples were filtered to include at least 500,000 reads (Supplementary Note 9). The output was aggregated to the individual clades using R78 v.4.4.1 and the tidyverse93 v.2.0.0 package. A full overview of all clades without aggregation is shown in Supplementary Fig. 9. Read abundance was normalized by HMM-alignment length (bacterial AmoA: 744 nucleotides, archaeal AmoA: 648 nucleotides, pNXR: 3,411 nucleotides, cNXR: 4,092 nucleotides) reported in RPKM. To reduce the complexity and describe the habitats with interesting nitrifier distributions, only the major soil types from MFDO1 habitats ‘soil, agriculture, fields’, ‘soil, natural, bogs, mires and fens’, ‘soil, natural, forests’, ‘soil, natural, grassland formations’ and ‘soil, urban, greenspaces’ were included in the main analysis. Agricultural samples from reclaimed lowlands were included in the ‘soil, agriculture, fields’ category. Analysis of the remaining habitats is provided in Supplementary Note 9. Read abundances were Hellinger-transformed using vegan94 v.2.6-6.1 (function decostand) before calculation of Bray–Curtis dissimilarity with the package parallelDist109 v.0.2.6 (function parDist) and performing the principal coordinate decomposition with the ape114 v.5.8 package (function pcoa). Samples were clustered within MFDO2 habitats using hierarchical clustering with the stats78 v.4.4.1 package (function hclust, method=ward.D2). As the MFDO2 for samples within ‘soil, natural, forests’ was not descriptive, being either ‘temperate forests’ or ‘forest (non-habitattype)’, the MFDO3 was used in the clustering of forest samples. A least squared distance linear model was used to calculate the linear correlation between RPKM of Nitrospira clade B amoA (dependent variable) and Palsa-1315 nxrA (independent variable) using the stats78 v.4.4.1 package (function lm). Spearman’s rank correlation was calculated using stats v.4.4.1 package (function cor.test).
Linkage of AOA MAGs to the major terrestrial amoA clades
The nucleotide sequences of all full-length amoA genes encoded in the 890 AOA genomes in the GlobDB155 (release 220) were exported using anvi’o145 (v8 development version). These 679 amoA sequences were combined with the 1,206 amoA sequences in the dataset from ref. 46 and aligned with Muscle5. The alignment was trimmed by removing the first 128 and last 41 positions using TrimAl151 v.1.5.0 to only include the 591 aligned positions used to create the phylogeny of ref. 46. A phylogeny was calculated based on the trimmed alignment with IQ-TREE152 v.2.4.0 using the GTR + F + I + R10 model selected by ModelFinder153. The GTDB R220 taxonomy of the GlobDB genomes encoding amoA sequences was matched to the amoA clades by manually inspecting the phylogeny. For the terrestrial Nitrososphaeraceae, the genera assigned by GTDB-Tk132 v.2.4.0 directly corresponded to the specific clades in the amoA phylogeny defined previously46. A translation table for the amoA and GTDB taxonomies of the Nitrososphaeraceae is provided in Supplementary Data 3. For the Nitrosopumilaceae (including the Nitrosotalea genus), a direct translation between amoA clades and GTDB genera could not be made for all clades defined in either amoA or concatenated marker gene phylogenies due to incongruencies in the topologies of the amoA and concatenated marker gene phylogenies.
Phylogenomic investigation of nitrifier genera, and Nitrobacter-like NxrA containing MAGs compared to NxrA gene phylogeny
We identified MAGs encoding Nitrobacter-like NxrA sequences of at least 600 amino acids in length within the family Xanthobacteraceae genera BOG-931, Bradyrhizobium, JAFAXD01, Pseudolabrys and VAZQ01. The MAGs encoding Nitrobacter-like NxrA sequences were selected for phylogenetic analysis, alongside species that belonged to the genus level GTDB34 R214 groups BOG-931, Bradyrhizobium, JAFAXD01, Pseudolabrys and VAZQ01, and also met the >90% genome completeness and <5% genome contamination thresholds using CheckM2 (ref. 131) v.1.0.2. The single recovered Nitrobacter MAG from this study was also included, as well as representatives from GTDB34 R214 for context, for example, the species representatives present for BOG-931, JAFAXD01, VAZQ01 and manually selected isolates from Bradyrhizobium and Pseudolabrys. The specific Nxr groups were assigned based on the updated GraftM package and classification tree. The phylogenomic tree was constructed using GTDB-Tk132 v.2.3.2 and the de_novo_wf on all bacterial MAGs. The multiple-sequence alignments of 120 single-copy proteins were subset to the genomes of interest using fxtract156 v.2.3. This alignment was used as input for IQ-TREE152 v.2.1.2 using the WAG + G model and -B 1000 using UFBoot. The tree was visualized in ARB102 v.7.0 for rerooting by the Methylopilaceae isolate outgroups and analysis, and further processed in iTOL154 v.6.
The cytoplasmic NxrA tree was built using Nitrobacter-like NxrA sequences of at least 600 amino acids in length from Xanthobacteraceae-family MAGs, along with known NxrA from Nitrococcus and Nitrobacter, and outgroup NxrA sequences from Nitrospira, Scalindua and Brocadia. Alignment, trimming and tree-generation was done using MAFFT150 v.7.490, TrimAl151 v.1.4.1, IQ-TREE152 v.2.2.0.3 with ultrafast bootstrap approximation and 1,000 iterations, using substitution model LG + F + R10.
Phylogenomic trees for known nitrifier genera as well as the putative genera examined in this study were created by subsetting the GTDB-Tk de_novo_wf trees for the bacteria and archaea. Trees were investigated and refined using ARB102 v.7.0 and iTOL v.6 as above, with final refinements in Inkscape v.1.4.2.
Metabolic investigation of nitrifier MAGs
Metabolic reconstruction of the short-read MAGs was conducted using DRAM157 v.1.4.6 and the KEGG database158 release 109. DRAM.py annotate was run using the default settings, followed by DRAM.py distil. Key nitrification and Calvin–Benson–Bassham-cycle genes were searched based on KO identifiers in the DRAM annotations.tsv output file and incorporated into the genome trees. Gene synteny was displayed using R78 v.4.4.1 and the gggenes111 v.0.5.1 package. To annotate carbohydrate active enzymes, dbCAN HMMdb159 v.13 (released 14 August 2024) was used on the dbCAN3 webserver, and hits were filtered (E < 10−15, coverage >0.80) as described previously48. Peptidases were identified with DRAM based on the MEROPS peptidase database160 (downloaded 1 July 2024) and were filtered to not include any hits to unassigned peptidases or non-peptidase homologues.
Data exploration and visualization
Data analysis was conducted in RStudio 2024.04.2 and 2023.12 using R78 v.4.2.3 to v.4.4.0 using tidyverse93 v.2.0.0. Data were read with either data.table161 v.1.15.4 or readxl162 v.1.4.3. Summary statistics are either reported as median or as mean ± s.d. where applicable. The colour scale for the MFOD1 levels of the ontology was made using wesanderson163 v.0.3.7. To ensure that continuous gradients were colour blind friendly, colours from viridisLite164 v.0.4.2 were used. Plots were made using ggplot from tidyverse93 v.2.0.0, patchwork165 v.1.2.0, ggpmisc166 v.0.5.6, ggpubr167 v.0.6.0 and ggtext168 v.0.1.2 and combined using Adobe Illustrator 2024 and Inkscape v.1.4.2.
Sample permissions
All necessary permits for sample collection were obtained before fieldwork. All samples were collected within the Danish Exclusive Economic Zone (EEZ); therefore, the Nagoya Protocol on Access and Benefit-Sharing does not apply. The Danish EEZ does not include any indigenous territories; accordingly, the CARE (collective benefit, authority to control, responsibility, and ethics) principles were not applicable to this work.
Etymology
Description of ‘Candidatus Nitronatura plena’ gen. nov., sp. nov.:
‘Candidatus Nitronatura plena’ (ni.tro.na.tu’ra. From L. neut. n. nitrum, nitrate, and N.L. fem. n. natura, nature; N.L. fem. n. Nitronatura, meaning ‘nitrogen and nature’, symbolizing a lineage associated with nitrogen cycling in natural ecosystems; ple’na. L. fem. adj. plena, full or complete, referring to the capacity for complete ammonia oxidation by clade B comammox). This taxon is represented by the circular closed Nitrospiraceae MAG GCA_974707355.1, recovered from a sphagnum acid bog. The complete protologue is provided in Supplementary Data 8.
Description of ‘Candidatus Nitrososappho danica’ gen. nov., sp. nov.:
‘Candidatus Nitrososappho danica’ (ni.tro.so.sap’pho. From L. masc. adj, nitrosus, full of natron, here intended to mean ‘nitrous’; and N.L. fem. n. Sappho, latinized form of the Greek poet’s name Σαπφώ. N.L. fem. n. Nitrososappho, is used here as a metaphor for resilience and persistence, in this case, of an archaeal ammonia oxidizer found in environmentally disturbed or constrained habitats; da’ni.ca. N.L. fem. adj. danica, Danish, referring to the origin of the sample and wide distribution in Denmark). This taxon is represented by the archaeal MAG (comprising 8 contigs) GCA_974504955.1, recovered from the sediment of an urban rainwater basin; however, the species is also widely present in agriculture. The complete protologue is provided in Supplementary Data 8.
The proposed names were registered at SeqCode under the register list seqco.de/r:xueh5m88.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data in NCBI are accessible through BioProject accession PRJNA1071982. This includes the sequencing data: bacterial UMI 16S rRNA gene reads (PromethION), bacterial rRNA operon data (PACBIO), eukaryotic rRNA operon (PACBIO) and metagenomes (Illumina), which are available at the NCBI SRA. Moreover, the high-quality MAGs, based on completeness and contamination (>90% completeness <5% contamination), are available at the NCBI GenBank under BioProject PRJNA1071982. The long-read MAG data are in the ENA under BioProject PRJEB586. Data supporting the findings of this study are available at Zenodo169 (https://doi.org/10.5281/zenodo.17162544). These data include the medium-quality MAGs, metagenome assemblies and MFG 16S rRNA gene reference database (clustered and unclustered). We also uploaded the amplicon data (UMI 16S rRNA gene, 16S from operons and 18S from operons) to the Global Biodiversity Information Facility as separate datasets which can be accessed online (https://doi.org/10.15468/33qqsm, https://doi.org/10.15468/yr5rmw and https://doi.org/10.15468/ea7jvq, respectively). Data (metadata and data files) at GitHub are accessible at https://github.com/cmc-aau/mfd_wiki/wiki. Public data used in this study include GTDB r220 and r214 (https://data.gtdb.ecogenomic.org/releases/), SingleM GTDB R220 reference metapackage v.4.3.0 (https://zenodo.org/records/11323477)139, SILVA v.138.1 (https://www.arb-silva.de/documentation/release-1381/), PR2 database v.5.0.0 (https://github.com/pr2database/pr2database/releases), MiDAS 4 and 5 databases (https://www.midasfieldguide.org/guide/downloads), AGP70 database (ENA: PRJEB32674; https://www.ebi.ac.uk/ena/browser/view/PRJEB32674)21; NCBI (SRA: PRJNA787301; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA787301/) MIDORI2 GB257 database (https://www.reference-midori.info/download.php), Greengenes2 (https://ftp.microbio.me/greengenes_release/2022.10/), GEM database (https://genome.jgi.doe.gov/portal/GEMs/GEMs.home.html), GlobDB release 220 (https://globdb.org/downloads), KEGG database release 109 (https://www.kegg.jp/kegg/docs/relnote.html), dbCAN HMMdb and dbCAN3 server (https://bcb.unl.edu/dbCAN2/), MEROPS Peptidase Database (https://www.ebi.ac.uk/merops/) and UniRef100 database (https://www.uniprot.org/uniref).
Code availability
The code for analyses and figure generation is available in GitHub (https://github.com/cmc-aau/mfd_wiki/wiki).
References
Sunagawa, S. et al. Tara Oceans: towards global ocean ecosystems biology. Nat. Rev. Microbiol. 18, 428–445 (2020).
Gacesa, R. et al. Environmental factors shaping the gut microbiome in a Dutch population. Nature 604, 732–739 (2022).
Shaffer, J. P. et al. Standardized multi-omics of Earth’s microbiomes reveals microbial and metabolite diversity. Nat. Microbiol. 7, 2128–2150 (2022).
Nayfach, S. et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 39, 499–509 (2021).
Schmidt, T. S. B. et al. SPIRE: a searchable, planetary-scale microbiome REsource. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad943 (2023).
Anker, J. From the early history of the Flora Danica. Libri 1, 334–350 (1950).
Thompson, L. R. et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017).
Directive, H. et al. Council directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora. Off. J. Eur. Union 206, 50 (1992).
Davies, C. E. & Moss, D. EUNIS Habitat Classification. Final report to the European Topic Centre on Nature Conservation (European Environment Agency, 1999).
Levin, G. Basemap04: Documentation of the Data and Method for the Elaboration of a Land Use and Land Cover Map for Denmark (Aarhus Univ., 2022); pure.au.dk/portal/en/publications/basemap04-documentation-of-the-data-and-method-for-the-elaboratio.
Karst, S. M. et al. High-accuracy long-read amplicon sequences using unique molecular identifiers with Nanopore or PacBio sequencing. Nat. Methods 18, 165–169 (2021).
Yarza, P. et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014).
Roswell, M., Dushoff, J. & Winfree, R. A conceptual guide to measuring species diversity. Oikos 130, 321–338 (2021).
Jost, L. Entropy and diversity. Oikos 113, 363–375 (2006).
Chiu, C.-H. & Chao, A. Estimating and comparing microbial diversity in the presence of sequencing errors. PeerJ 4, e1634 (2016).
Guillou, L. et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604 (2013).
Edgar, R. C. Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ 6, e4652 (2018).
Dueholm, M. S. et al. Generation of comprehensive ecosystem-specific reference databases with species-level resolution by high-throughput full-length 16S rRNA gene sequencing and automated taxonomy assignment (AutoTax). mBio 11, e01557-20 (2020).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Dueholm, M. K. D. et al. MiDAS 5: global diversity of bacteria and archaea in anaerobic digesters. Nat. Commun. 15, 5361 (2024).
Overgaard, C. K. et al. Application of ecosystem-specific reference databases for increased taxonomic resolution in soil microbial profiling. Front. Microbiol. 13, 942396 (2022).
McDonald, D. et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01845-1 (2023).
Louca, S., Mazel, F., Doebeli, M. & Parfrey, L. W. A census-based estimate of Earth’s bacterial and archaeal diversity. PLoS Biol. 17, e3000106 (2019).
Walters, K. E. & Martiny, J. B. H. Alpha-, beta-, and gamma-diversity of bacteria varies across habitats. PLoS ONE 15, e0233872 (2020).
Labouyrie, M. et al. Patterns in soil microbial diversity across Europe. Nat. Commun. 14, 3311 (2023).
Delgado-Baquerizo, M. et al. Global homogenization of the structure and function in the soil microbiome of urban greenspaces. Sci. Adv. 7, eabg5809 (2021).
Peng, Z. et al. Land conversion to agriculture induces taxonomic homogenization of soil microbial communities globally. Nat. Commun. 15, 3624 (2024).
Foley, J. A. et al. Global consequences of land use. Science 309, 570–574 (2005).
Urbanová, Z. & Bárta, J. Microbial community composition and in silico predicted metabolic potential reflect biogeochemical gradients between distinct peatland types. FEMS Microbiol. Ecol. 90, 633–646 (2014).
Langendries, S. & Goormachtig, S. Paenibacillus polymyxa, a Jack of all trades. Environ. Microbiol. 23, 5659–5669 (2021).
Brunbjerg, A. K. et al. A systematic survey of regional multi-taxon biodiversity: evaluating strategies and coverage. BMC Ecol. 19, 43 (2019).
Neu, A. T., Allen, E. E. & Roy, K. Defining and quantifying the core microbiome: challenges and prospects. Proc. Natl Acad. Sci. USA 118, e2104429118 (2021).
O’Malley, M. A. “Everything is everywhere: but the environment selects”: ubiquitous distribution and ecological determinism in microbial biogeography. Stud. Hist. Philos. Biol. Biomed. Sci. 39, 314–325 (2008).
Parks, D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794 (2022).
Petersen, R. J., Blicher-Mathiesen, G., Rolighed, J., Andersen, H. E. & Kronvang, B. Three decades of regulation of agricultural nitrogen losses: experiences from the Danish Agricultural Monitoring Program. Sci. Total Environ. 787, 147619 (2021).
Beeckman, F., Motte, H. & Beeckman, T. Nitrification in agricultural soils: impact, actors and mitigation. Curr. Opin. Biotechnol. 50, 166–173 (2018).
Petersen, S. O. et al. Beskrivelse af Dokumentationsindsats Vedr. Klimaeffekter, Opdatering af Aktivitetsdata og Afdækning af Væsentlige Sideeffekter for Anvendelsen af Syntetiske Nitrifikationshæmmere (Aarhus Univ., 2025); pure.au.dk/ws/portalfiles/portal/426309762/Levering_dokumentationsindsats_syntetiske_nitrifikationsh_mmere.pdf.
Rojas-Pinzon, P. A. et al. Inhibition profile of three biological nitrification inhibitors and their response to soil pH modification in two contrasting soils. FEMS Microbiol. Ecol. 100, fiae072 (2024).
Boyd, J. A., Woodcroft, B. J. & Tyson, G. W. GraftM: a tool for scalable, phylogenetically informed classification of genes within metagenomes. Nucleic Acids Res. 46, e59 (2018).
Woodcroft, B. J. et al. Comprehensive taxonomic identification of microbial species in metagenomic data using SingleM and Sandpiper. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02738-1 (2025).
Shaw, J. & Yu, Y. W. Rapid species-level metagenome profiling and containment estimation with sylph. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02412-y (2024).
Simon, J. & Klotz, M. G. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim. Biophys. Acta 1827, 114–135 (2013).
Prosser, J. I., Hink, L., Gubry-Rangin, C. & Nicol, G. W. Nitrous oxide production by ammonia oxidizers: physiological diversity, niche differentiation and potential mitigation strategies. Glob. Change Biol. 26, 103–118 (2020).
Huang, L. et al. Ammonia-oxidizing archaea are integral to nitrogen cycling in a highly fertile agricultural soil. ISME Commun. 1, 19 (2021).
Jia, Z. & Conrad, R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11, 1658–1671 (2009).
Alves, R. J. E., Minh, B. Q., Urich, T., von Haeseler, A. & Schleper, C. Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes. Nat. Commun. 9, 1517 (2018).
Könneke, M. et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc. Natl Acad. Sci. USA 111, 8239–8244 (2014).
Bei, Q. et al. Metabolic potential of Nitrososphaera-associated clades. ISME J. 18, wrae086 (2024).
Alves, R. J. E. et al. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J. 7, 1620–1631 (2013).
Hu, J. et al. Dominance of comammox Nitrospira in soil nitrification. Sci. Total Environ. 780, 146558 (2021).
Li, D., Fang, F. & Liu, G. Efficient nitrification and low N2O emission in a weakly acidic bioreactor at low dissolved oxygen levels are due to comammox. Appl. Environ. Microbiol. 87, e00154-21 (2021).
Daims, H. et al. Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 (2015).
Woodcroft, B. J. et al. Genome-centric view of carbon processing in thawing permafrost. Nature 560, 49–54 (2018).
Greco, C., Andersen, D. T., Yallop, M. L., Barker, G. & Jungblut, A. D. Genome-resolved metagenomics reveals diverse taxa and metabolic complexity in Antarctic lake microbial structures. Environ. Microbiol. 26, e16663 (2024).
Li, C., Hu, H.-W., Chen, Q.-L., Chen, D. & He, J.-Z. Niche differentiation of clade A comammox Nitrospira and canonical ammonia oxidizers in selected forest soils. Soil Biol. Biochem. 149, 107925 (2020).
Yuan, D. et al. Comammox activity dominates nitrification process in the sediments of plateau wetland. Water Res. 206, 117774 (2021).
Li, C., Hu, H.-W., Chen, Q.-L., Chen, D. & He, J.-Z. Comammox Nitrospira play an active role in nitrification of agricultural soils amended with nitrogen fertilizers. Soil Biol. Biochem. 138, 107609 (2019).
Attard, E. et al. Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 12, 315–326 (2010).
Sereika, M. et al. Genome-resolved long-read sequencing expands known microbial diversity across terrestrial habitats. Nat. Microbiol. 10, 2018–2030 (2025).
Starkenburg, S. R. et al. Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl. Environ. Microbiol. 74, 2852–2863 (2008).
Wertz, S., Leigh, A. K. K. & Grayston, S. J. Effects of long-term fertilization of forest soils on potential nitrification and on the abundance and community structure of ammonia oxidizers and nitrite oxidizers. FEMS Microbiol. Ecol. 79, 142–154 (2012).
Séneca, J. et al. Composition and activity of nitrifier communities in soil are unresponsive to elevated temperature and CO2, but strongly affected by drought. ISME J. 14, 3038–3053 (2020).
Edgar, R. C. SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. Preprint at bioRxiv https://doi.org/10.1101/074161 (2016).
Bruun, A.-M., Finster, K., Gunnlaugsson, H. P., Nørnberg, P. & Friedrich, M. W. A comprehensive investigation on iron cycling in a freshwater seep including microscopy, cultivation and molecular community analysis. Geomicrobiol. J. 27, 15–34 (2010).
Orgiazzi, A., Ballabio, C., Panagos, P., Jones, A. & Fernández-Ugalde, O. LUCAS Soil, the largest expandable soil dataset for Europe: a review. Eur. J. Soil Sci. 69, 140–153 (2018).
Frøslev, T. G. et al. The biodiversity effect of reduced tillage on soil microbiota. Ambio 51, 1022–1033 (2022).
Frøslev, T. G. et al. Treated like dirt: robust forensic and ecological inferences from soil eDNA after challenging sample storage. Environ. DNA 5, 158–174 (2023).
Haasler, S. et al. Recycling of phosphorus from dredged lake sediment: Importance of iron-bound phosphates for plant growth. Sustain. Environ. 10, 2362503 (2024).
Jensen, M. R. et al. The core of the matter-importance of identification method and biological replication for benthic marine monitoring. Ecol. Evol. 14, e70556 (2024).
Beulig, F., Røy, H., Glombitza, C. & Jørgensen, B. B. Control on rate and pathway of anaerobic organic carbon degradation in the seabed. Proc. Natl Acad. Sci. USA 115, 367–372 (2018).
Marshall, I. P. G. et al. Environmental filtering determines family-level structure of sulfate-reducing microbial communities in subsurface marine sediments. ISME J. 13, 1920–1932 (2019).
Beulig, F., Røy, H., McGlynn, S. E. & Jørgensen, B. B. Cryptic CH4 cycling in the sulfate–methane transition of marine sediments apparently mediated by ANME-1 archaea. ISME J. 13, 250–262 (2019).
Maksimavičius, E. & Roslev, P. Methane emission and methanotrophic activity in groundwater-fed drinking water treatment plants. Water Sci. Technol. Water Supply 20, 819–827 (2020).
Palomo, A., Dechesne, A., Pedersen, A. G. & Smets, B. F. Genomic profiling of Nitrospira species reveals ecological success of comammox Nitrospira. Microbiome 10, 204 (2022).
Dottorini, G. et al. Mass-immigration determines the assembly of activated sludge microbial communities. Proc. Natl Acad. Sci. USA 118, e2021589118 (2021).
Breda, I. L., Ramsay, L., Søborg, D. A., Dimitrova, R. & Roslev, P. Manganese removal processes at 10 groundwater fed full-scale drinking water treatment plants. Water Qual. Res. J. Can. 54, 326–337 (2019).
Jensen, T. B. N., Dall, S. M., Knutsson, S., Karst, S. M. & Albertsen, M. High-throughput DNA extraction and cost-effective miniaturized metagenome and amplicon library preparation of soil samples for DNA sequencing. PLoS ONE 19, e0301446 (2024).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018).
Grolemund, G. & Wickham, H. Dates and times made easy with Lubridate. J. Stat. Softw. 40, 1–25 (2011).
Hijmans, R. J. Terra: spatial data analysis. R package version 1.7-78. CRAN https://cran.r-project.org/web/packages/terra/index.html (2024).
EEA Geospatial Data Catalogue 1 km (EEA, 2013).
EEA Geospatial Data Catalogue 10 km (EEA, 2013).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Tange, O. GNU Parallel 20220722 (‘Roe vs Wade’). Zenodo https://doi.org/10.5281/zenodo.6891516 (2022).
Adler, M. Pigz. GitHub https://github.com/madler/pigz (2017).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013).
Pruesse, E., Peplies, J. & Glöckner, F. O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012).
Cavalier-Smith, T., Lewis, R., Chao, E. E., Oates, B. & Bass, D. Helkesimastix marina n. sp. (Cercozoa: Sainouroidea superfam. n.) a gliding zooflagellate of novel ultrastructure and unusual ciliary behaviour. Protist 160, 452–479 (2009).
López-García, P., Philippe, H., Gail, F. & Moreira, D. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl Acad. Sci. USA 100, 697–702 (2003).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019).
Oksanen, J. et al. Vegan: community ecology package. R package version 2.6-6.1. CRAN https://cran.r-project.org/web/packages/vegan/vegan.pdf (2024).
Hsieh, T. C., Ma, K. H. & Chao, A. iNEXT: an R package for rarefaction and extrapolation ofspecies diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456 (2016).
Kassambara, A. Rstatix: pipe-friendly framework for basic statistical tests. R package version 0.7.2. CRAN https://cran.r-project.org/web/packages/rstatix/rstatix.pdf (2023).
Dueholm, M. K. D. et al. MiDAS 4: a global catalogue of full-length 16S rRNA gene sequences and taxonomy for studies of bacterial communities in wastewater treatment plants. Nat. Commun. 13, 1908 (2022).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Lane, D. J. in Nucleic Acid Techniques in Bacterial Systematics (ed. Goodfellow, E. S. A.) 205–248 (John Wiley & Sons, 1991).
Massana, R., Murray, A. E., Preston, C. M. & DeLong, E. F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63, 50–56 (1997).
Bahram, M., Anslan, S., Hildebrand, F., Bork, P. & Tedersoo, L. Newly designed 16S rRNA metabarcoding primers amplify diverse and novel archaeal taxa from the environment. Environ. Microbiol. Rep. 11, 487–494 (2019).
Ludwig, W. et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004).
Leray, M., Knowlton, N. & Machida, R. J. MIDORI2: A collection of quality controlled, preformatted, and regularly updated reference databases for taxonomic assignment of eukaryotic mitochondrial sequences. Environ. DNA 4, 894–907 (2022).
Kalvari, I. et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49, D192–D200 (2021).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
Article MathSciNet CAS PubMed PubMed Central ADS Google Scholar
Wheeler, T. J. & Eddy, S. R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489 (2013).
Guenard, G., Legendre, P. & Pages, B. Codep: multiscale codependence analysis. R package version 1.2-3. CRAN https://cran.r-project.org/web/packages/codep/codep.pdf (2025).
Andersen, K. S., Kirkegaard, R. H., Karst, S. M. & Albertsen, M. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. Preprint at bioRxiv https://doi.org/10.1101/299537 (2018).
Eckert, A. ParallelDist: parallel distance matrix computation using multiple threads. R package version 0.2.4. CRAN https://cran.r-project.org/web/packages/parallelDist/parallelDist.pdf (2018).
Carr, D., Lewin-Koh, N., Maechler, M., Sarkar, D. & Pebesma, E. Hexagonal binning routines. CRAN https://cran.r-project.org/package=hexbin (2024).
Wickham, H. Ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Dunnington, D., Thorne, B. & Hernangómez, D. Spatial data framework for ggplot2. R package version 1.1.9. CRAN https://cran.r-project.org/package=ggspatial (2023).
Massicotte, P., South, A. & Hufkens, K. World map data from Natural Earth. R package version 1.0.1. CRAN https://cran.r-project.org/package=rnaturalearth (2023).
Emmanuel Paradis, S. B. et al. Ape: analyses of phylogenetics and evolution. R package version 5.8. CRAN https://cran.r-project.org/web/packages/ape/index.html (2024).
Graves, S., Piepho, H.-P., Selzer, L. & Dorai-Raj, S. multcompView: visualizations of paired comparisons v0.1-10. CRAN https://cran.r-project.org/web/packages/multcompView/index.html (2024).
Gibb, S. bootstrap: simple bootstrapping for hierarchical clustering. R package version 0.1. GitHub https://codeberg.org/sgibb/bootstrap (2013).
Naimi, B. & Araújo, M. B. sdm: a reproducible and extensible R platform for species distribution modelling. Ecography 39, 368–375 (2016).
Leonardi, M., Colucci, M., Pozzi, A., Scerri, E. & Manica, A. tidysdm: species distribution models with Tidymodels. R package version 0.9.5. GitHub https://github.com/EvolEcolGroup/tidysdm (2023).
Kuhn, M., Wickham, H. & Posit Software. Tidymodels: easily install and load the ‘Tidymodels’ packages. R package version 1.2.0. CRAN https://cran.r-project.org/web/packages/tidymodels/tidymodels.pdf (2024).
Hvitfeldt, E. Themis: extra recipes steps for dealing with unbalanced data. R package version 1.0.3. tidymodels https://themis.tidymodels.org (2025).
Wright, M. N., Wager, S. & Probst, P. Ranger: a fast implementation of random forests. R package version 0.16.0. CRAN https://cran.r-project.org/web/packages/ranger/ranger.pdf (2023).
Kuhn, M., Vaughan, D., Hvitfeldt, E. & Posit Software. Yardstick: tidy characterizations of model performance. R package version 1.3.1. CRAN https://cran.r-project.org/web/packages/yardstick/index.html (2024).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Krassowski, M. ComplexUpset: create complex UpSet plots using ‘ggplot2’ components. R package version 1.3.3. CRAN https://cran.r-project.org/web/packages/ComplexUpset/ComplexUpset.pdf (2021).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Woodcroft, B. J. Default SingleM reference “metapackage” data. Zenodo https://doi.org/10.5281/zenodo.5739611 (2023).
Aroney, S. T. N., Newell, R. J. P., Tyson, G. W. & Woodcroft, B. J. Bin Chicken: targeted metagenomic coassembly for the efficient recovery of novel genomes. Nat. Methods https://doi.org/10.1038/s41592-025-02901-1 (2025).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019).
Chklovski, A., Parks, D. H., Woodcroft, B. J. & Tyson, G. W. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 20, 1203–1212 (2023).
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019).
Schwengers, O. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 7, 000685 (2021).
Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49, 9077–9096 (2021).
Seemann, T. Barrnap. GitHub https://github.com/tseemann/barrnap (2018).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017).
Aroney, S. T. N. et al. CoverM: read alignment statistics for metagenomics. Bioinformatics 41, btaf147 (2025).
Woodcroft, B. J. Default SingleM reference ‘metapackage’ data. Zenodo https://doi.org/10.5281/zenodo.11323477 (2024).
Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Suzek, B. E., Huang, H., McGarvey, P., Mazumder, R. & Wu, C. H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288 (2007).
Shaw, J., Marin, M. G. & Li, H. High-resolution metagenome assembly for modern long reads with myloasm. Preprint at bioRxiv https://doi.org/10.1101/2025.09.05.674543 (2025).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T. & Aluru, S. High-throughput ANI analysis of 90 K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018).
Eren, A. M. et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 6, 3–6 (2021).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Rasko, D. A., Myers, G. S. A. & Ravel, J. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinform. 6, 2 (2005).
Speth, D. R. & Orphan, V. J. Metabolic marker gene mining provides insight in global mcrA diversity and, coupled with targeted genome reconstruction, sheds further light on metabolic potential of the Methanomassiliicoccales. PeerJ 6, e5614 (2018).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Speth, D. R. et al. GlobDB: a comprehensive species-dereplicated microbial genome resource. Bioinform. Adv. 5, vbaf280 (2025)
Skennerton, C. T. Fxtract: extract sequences from a Fastx file given a subsequence or identifier. GitHub https://github.com/ctSkennerton/fxtract (2021).
Shaffer, M. et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 48, 8883–8900 (2020).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Zheng, J. et al. dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 51, W115–W121 (2023).
Rawlings, N. D. et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 46, D624–D632 (2018).
Barrett, T. et al. data.table: extension of ‘data.frame’. R package version 1.15.4. CRAN https://cran.r-project.org/package=data.table (2024).
Wickham, H. & Bryan, J. readxl: read excel files. R package version 1.4.3. CRAN https://cran.r-project.org/package=readxl (2023).
Ram, K., Wickham, H., Richards, C. & Aaron, B. wesanderson: a Wes Anderson palette generator. R package version 0.3.7. CRAN https://cran.r-project.org/package=wesanderson (2023).
Garnier, S. viridisLite: colorblind-friendly color maps (lite version). R package version 0.4.2. CRAN https://cran.r-project.org/package=viridisLite (2023).
Pedersen, T. L. patchwork: the composer of plots. R package version 1.2.0. CRAN https://cran.r-project.org/package=patchwork (2024).
Aphalo, P. J. ggpmisc: miscellaneous extensions to ‘ggplot2’. R package version 0.6.2. CRAN https://cran.r-project.org/package=ggpmisc (2025).
Kassambara, A. ggpubr: ‘ggplot2’ based publication ready plots. R package version 0.6.0. CRAN https://cran.r-project.org/package=ggpubr (2023).
Wilke, C. O. & Wiernik, B. M. ggtext: improved text rendering support for ‘ggplot2’. R package version 0.1.2. CRAN https://cran.r-project.org/package=ggtext (2022).
Singleton, C. M. et al. Data for ‘Microflora Danica: the atlas of Danish environmental microbiomes’. Zenodo https://doi.org/10.5281/zenodo.17162544 (2025).
Acknowledgements
Funding was provided by the Poul Due Jensen Foundation, PDJF (grant MicroFlora Danica to M.A. and P.H.N.), Villum Foundation (grant 15510 and 50093 to M.A., grant 13351 to P.H.N.), the European Union (ERC grant 101078234 to M.A.), the Velux Foundation (grant 21517 to P.F.T.) and the Carlsberg Foundation (grant CF18-0949 to P.F.T.). C.M.S. was supported by a Novo Nordisk Foundation Postdoctoral Fellowship grant (NNF20OC0065005); D.R.S. and M.W. by the Cluster of Excellence grant ‘Microbiomes drive Planetary Health’ of the Austrian Science Fund FWF (10.55776/COE7); S.T.N.A. by the EMERGE National Science Foundation (NSF) Biology Integration Institute (2022070); B.J.W. by an Australian Research Council Future Fellowship (FT210100521). This study is funded by the European Union Horizon Europe research and innovation program under grant agreement no. 101086179 (AI4SoilHealth). Views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the Research Executive Agency. Neither the European Union nor the granting authority can be held responsible for them. We thank the members of the Microflora Danica Consortium for their contributions in collecting samples and related metadata across Denmark.
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Brett Baker and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Distribution of the samples per MFDO1 habitats in the 10 km EU reference grid of Denmark.
The map of Denmark follows the same criteria and scales as Fig. 1a, including the cutout for Bornholm island, with the base map from Eurostat (Geodata - GISCO - Eurostat). Each facet includes only samples of the relevant MFDO1 category as indicated in each title. Most MFDO1 habitats (18, representing 9,975 samples) exhibited broad geographical sample distributions across the country. An exception was MFDO1 “Rocky habitats and caves”, with samples only found on the island of Bornholm, which is geologically different from the rest of Denmark. The 10 km EU reference grid of Denmark was overlaid to the map and each cell where at least one metagenomic sample was present has been shaded. Cells including at least one sample with amplicon data have been marked with a dot in the centre.
Extended Data Fig. 2 Eukaryotic diversity of the Danish habitats.
a. Diversity overview of the selected habitats with each facet addressing a different measure of diversity. The selected 10 MFDO1 habitats are represented in the rows of the multi-facet plot. The n = X number of biologically independent samples per habitat is indicated by the # FL18S column. Dendrogram of between-group (branches) and within-group (nodes) Jaccard dissimilarity. Bootstrap values were calculated using 100 iterations. A heatmap of the relative abundances of the 20 most prevalent eukaryotic Divisions-Subdivisions are shown, taxonomy is inferred with the PR2 database v5.0.0. The three hinges of the box plots correspond to 25th, 50th and 75th percentiles of the distributions whilst the whiskers extend to a maximum of 1.5 times the distance between the 25th and 75th percentile hinges. All individual samples are shown as points (with a jitter to improve visualization). Gamma diversity (Hill-Shannon diversity) is reported with corresponding error bars spanning the 5th-95th percentiles of the distribution. b. Ordination of the 18S rRNA dataset. 18S rRNA sequences from eukaryotic rRNA operon sequencing of 363 representative samples are arranged using PCoA and coloured according to MFDO1 habitat description together with the results from the ANOSIM and PERMANOVA. The visualization depicts the first two components, accounting for 3.3% and 2% of explained variance, respectively. c. Sub-panels of the individual 10 selected MFDO1 habitats are highlighted in different panels and colours together with the results of the contrasts analysis. The samples of each habitat often occupy a distinct portion (of variable size) of the plot.
Extended Data Fig. 3 Distance decay analysis.
The analysis was conducted on genus-aggregated 16S rRNA data subjected to random subsampling without replacement (1,002) reads. The samples were filtered to the samples with reliable coordinates (except from GPS masked “Agricultural” samples) and limited to distances below 300 km, excluding samples from Bornholm. This left 9,121 samples (9,132,242 reads) for the calculation of Hellinger-transformed Bray-Curtis and geographical distances (a total of 184,844,898 comparisons). The influence of spatial distance on the microbial community in the metagenome samples was negligible, except at short distances (< 10 km). In the top panel, points represent mean Bray-Curtis similarity of 5 km bins and error bars span 1 standard deviation.
Extended Data Fig. 4 Core genera across MFDO1 habitats.
a. Upset plot of unique and uniquely shared core genera identified across the 19 MFDO1 habitats. b. The prevalence and mean abundance of selected core genera within the families Nitrosomonadaceae, Nitrososphaeraceae and Nitrospiraceae across the MFDO1 soil habitats in the geographically balanced dataset. *Historically putative nitrifier groups, but capability is unknown. c. Stacked bar plot of the count and the cumulative relative abundance of the identified core and non-core genera as well as the proportion of reads unclassified at the genus level across the investigated MFDO1 habitats.
Extended Data Fig. 5 Novelty of Danish microbes.
a. The known species fraction of the microbial community in each metagenome estimated using SingleM against GTDB R220 (unshaded, see Methods) and with the MFD MAGs included (grey-shaded). The first four boxes represent 81,709 public metagenomes downloaded from NCBI SRA, with human gut metagenomes as a reference point of a well-studied system. MD indicates the median percentage of known species fraction per habitat. The two boxplots per habitat type represent the known species fraction when classifying using the SingleM database with MFD MAGs (+ MFD MAGs) and without MFD MAGs (-MFD MAGs). b. Basic MAG recovery statistics and taxonomic novelty against GTDB R220. The rows of the table line up with the habitats described in a. c. Boxplot of the proportions of the prokaryotic community represented by MAGs reconstructed from short-read metagenomes across different habitats. MD: median, sd: standard deviation. The sample number of the analyses, N/n = X biologically independent samples is indicated for each box. The three hinges of the box plots correspond to 25th, 50th and 75th percentiles of the distributions whilst the whiskers extend to a maximum of 1.5 times the distance between the 25th and 75th percentile hinges. All individual samples are shown as points (with a jitter to improve visualization).
Extended Data Fig. 6 Genome-based quantification of Nitrososphaeraceae AOAs.
a. Heatmap displaying the relative “taxonomic abundance” quantified at the genome level by sylph41 of AOA (families Nitrososphaeraceae and Nitrosopumilaceae) MAGs recovered in the MFD project and aggregated into species clusters. The AOA abundances shown are relative to the taxonomic abundance of all MFD MAGs. The samples are faceted by MFDO1 habitat type and clustered within each MFDO2 habitat, displayed with a colour bar on the x-axis. On the y-axis, species clusters are shown with GTDB-Tk132 classification and a species cluster ID. In cases where members of the same species cluster have several GTDB-Tk classifications, the name describing the most MAGs in the species cluster was chosen as the species cluster name. b. Relative taxonomic abundance of the most abundant AOA species cluster TA-21 sp02254895 cluster 98_1 in agricultural samples. To balance the dataset and ensure that habitats were compared on a national, and not local scale, one sample was randomly selected from each 10 km reference cell within each MFDO3 level. Only MFDO3 levels represented in at least 10 different 10 km reference cells were included in the analysis, to exclude habitat types only present in a confined area of Denmark. Samples are coloured by the MFDO2, point jitter is added for visualization, and the mean of each group is indicated in red. Pairwise comparison was made between each group (MFDO3) against all (base mean) using a one-sided (greater) Mann-Whitney U test, p-value was adjusted for multiple comparisons using Benjamini-Hochberg approach and significance level is indicated above each group (* = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001). The dotted line represents the mean of all samples (m = 4.65). c. Relative taxonomic abundance of TA-21 sp02254895 cluster 98_1 in “Greenspaces”, “Grassland formations”, and “Forests” samples. One sample was randomly selected from each 10 km reference cell within each MFDO3 level. Only MFDO3 levels represented in at least 10 different 10 km reference cells were included in the analysis. Samples are coloured by the MFDO1, point jitter is added for visualization, and the mean of each group is indicated in red. Pairwise comparison was made between each group (MFDO3) against all (base mean) using a one-sided (less) Mann-Whitney U test, p-value was adjusted for multiple comparisons using Benjamini-Hochberg approach and significance level is indicated above each group (* = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001). The dotted line represents the mean of all samples (m = 0.46). The sample number of the analyses, n = X biologically independent samples, is indicated in the x-axis labels of the violin plots.
Extended Data Fig. 7 Correlation of gene read abundance of comammox clade B amoA and Palsa-1315 nxrA.
Relative gene read abundance in selected MFDO1 habitats ‘Agriculture’, ‘Bogs, mires and fens’, ‘Forests’, ‘Urban Greenspaces’, ‘Grassland formations’, and ‘Natural Sediment’. Read abundance is reported in reads per kilobase million [RPKM]. A regression line generated with a least squared distance linear model is displayed in red. The regression coefficient (Reg. coeff) and R2 were computed using the lm function (stats78 v4.4.1 package) and Spearman’s rank correlation (ρ) was computed using the cor.test function (stats v4.4.1 package). P-values were adjusted with the Benjamini-Hochberg approach. The sample number of the analyses, n = X biologically independent samples, is indicated in the subplots.
Extended Data Fig. 8 Aligned genome and protein tree of new Nitrobacter-like narG/nxrA encoding MAGs.
Genome tree (left, alignment length 5,035aa) of potential nitrite oxidizers containing Nitrobacter-like NxrA, belonging to the Xanthobacteraceae, primarily Bradyrhizobium spp., genus BOG-931 spp., and Pseudolabrys spp. Protein tree (right) of NxrA sequences (alignment length 1233aa) from Nitrococcus, Nitrobacter, and Nitrobacter-like NxrA recovered from short read MAGs (SR-MAGs). Wedges represent clusters of genomes within the genera that do not contain the NxrA-like sequences of interest. Protein subunits involved in nitrite oxidation or reduction are indicated by: NxrA/NarG, NxrX, NxrB/NarH, NarJ, NxrC/NarI; nitrite/nitrate transport: NarK/NasA; formate_nitrite_transporter, electron transfer: Cyt.c class I; nitrite reduction: NirK, NirS; nitric oxide reduction: NorB, NorC; nitrous oxide reduction: NosZ; carbon dioxide fixation: CbbL, CbbS, PrK; nitrogen fixation: NifD, NifK, NifH. Tree scale bars represent the number of amino acid substitutions per site. ITOL v6 was used for tree display.
Extended Data Fig. 9 Genome tree of BOG-931 short read and long read MAGs of high quality.
Genome tree (left) of potential nitrite oxidizers containing Nitrobacter-like narG/nxrA, belonging to the genus BOG-931. LR-MAGs (MFDxxxxx.bin.xx) included are of HQ based on the MIMAG136 standard, while SR-MAGs (LIB-MJxxx-xx) with completeness >90% and contamination <5% have been included. Genomes in red encode Nitrobacter-like nxrA sequences. For LR-MAGs, gene synteny is only displayed for contigs that also encode Nitrobacter-like nxrA. In the circular genome MFD09603.bin.c.1, RuBisCO is present on the same contig, with a distance between nxr/nar and RuBisCO gene operon of 1.4 Mbp. Other nitrification genes on the same contig as Nitrobacter-like NarG/cNXR are displayed. In the box (dotted line) are nxr/nar genes from SR-MAGs, along with RuBisCO genes. Here, genes are located on several contigs. The arrangement of genes from different contigs is done manually for visual comparability. Wedges represent clusters of genomes within the genera that do not contain the NxrA-like sequences of interest, displaying the number of genomes collapsed into each clade.
Supplementary information
Supplementary Information
Supplementary Notes 1–10, Supplementary Figs. 1–10 and Supplementary Tables 1–4, containing information supporting the findings of the study and describing assessment of methods.
Reporting Summary
Supplementary Data 1
All core genera identified across all levels of the MFD ontology. List of core genera identified across the five levels of the Microflora Danica ontology (sample type, area type, MFDO1, MFDO2 and MFDO3). The relative abundance of each genus is reported as median, mean and the s.d. of habitat class-level specific abundance. Besides the taxonomy from phylum to genus, the table contains n_obserservations, the number of samples within an ontology level and habitat a genera is observed in; n_abundant, the number of samples where the genera has ≥0.1% relative abundance; median, mean and s.d. of the relative abundance, the number of samples in the specific habitat; group_size, the number of samples in the habitat category; core, the number of samples the genus needs to have ≥0.1% relative abundance to be considered part of the core; prevalence, the fraction of samples where the genus has ≥0.1% relative abundance; and fidelity, how many habitats of the specific ontology level the genus is observed in.
Supplementary Data 2
The number of core genera across the MFD ontology levels. The number of core genera identified across the five levels of the MFD ontology (sample type, area type, MFDO1, MFDO2, MFDO3). The columns to the right of an ontology level refer to the number of core genera for that habitat ontology class level.
Supplementary Data 3
AOA classifications. Classification matches of AOA genome to amoA clade taxonomy. The table contains genome identifier and GTDB genome taxonomy from the phylum to the species level, presence of the amoA gene in the genome and the specific clades in the amoA phylogeny defined previously46.
Supplementary Data 4
Functional annotation of nitrifiers. DRAM functional annotation output (annotations.tsv file) for AOA, AOB and NOB described in this study.
Supplementary Data 5
Summary of the sampling and extraction methods. Overview of sample type, sampling methods and number of samples across the different sampling projects in Microflora Danica. Where indicated by an asterisk, the sampling protocol used was very similar to the one used for the Microflora Danica sampling campaign65.
Supplementary Data 6
All sample metadata including methodology. Full metadata file with information relating to sampling, extraction, library preparation and sequencing. A full explanation of all fields can be found at GitHub (https://github.com/cmc-aau/mfd_metadata). Where indicated by an asterisk, the sampling protocol used was very similar to the one used for the Microflora Danica sampling campaign65.
Supplementary Data 7
Primer sequences used for UMI 16S and operon sequencing and processing. Primer sequences and primer binding positions referenced in the main and in the methods.
Supplementary Data 8
Protologue tables for ‘Candidatus Nitronatura plena’ and ‘Candidatus Nitrososappho danica’.
Peer Review File
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Singleton, C.M., Jensen, T.B.N., Delogu, F. et al. The Microflora Danica atlas of Danish environmental microbiomes. Nature 649, 971–981 (2026). https://doi.org/10.1038/s41586-025-09794-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09794-2