
🚨#COVID19 PAPER ALERT🚨
F@h simulations of the #coronavirus Nsp16 protein (1) reveal the structural mechanism of its activation, which ultimately enables the virus to escape our immune system, & (2) detect a ‘cryptic’ pocket to block Nsp16’s action.
biorxiv.org/content/10.110…
For successful infection, #coronaviruses use a protein called Nsp16 that modifies the viral genome so that the virus avoids an immune response. Nsp16 works by binding 2 substrates: RNA (green) and SAM (pink). Viruses and humans contain similar (homologous) proteins. 
Unlike other viruses and humans, #coronavirus Nsp16 requires a binding partner (Nsp10) to activate its enzymatic activity. This difference is interesting from a basic science standpoint, and because it could present a way to target the coronavirus selectively with a drug. 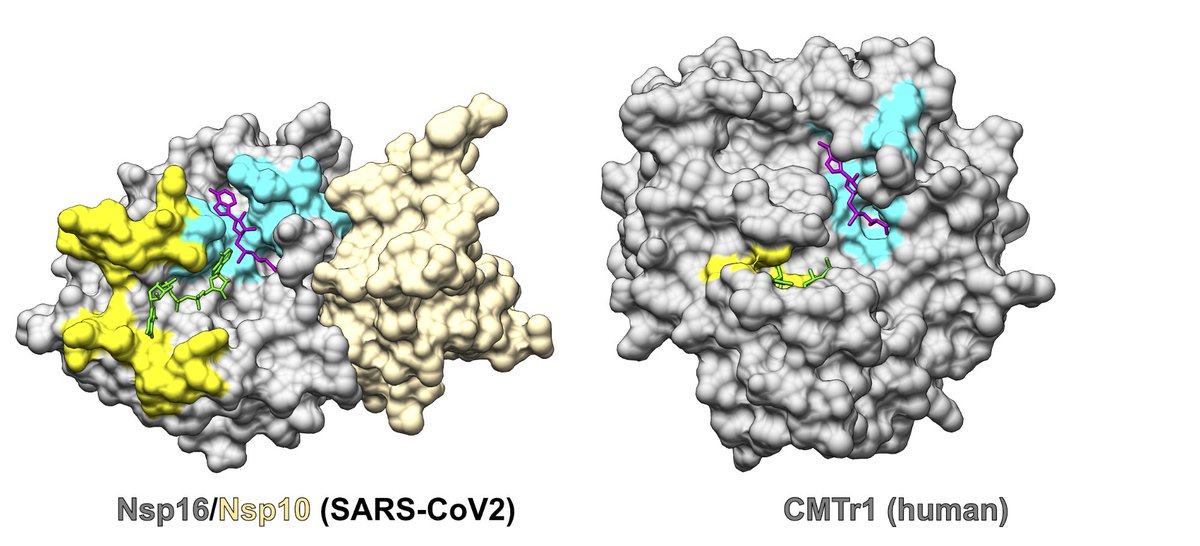
First, we wondered, how does Nsp10 activate Nsp16? We analyzed over 1.1 milliseconds of Folding@home simulations of Nsp16 with & without Nsp10 present and found striking structural differences using our recently developed deep learning tool (DiffNets) and Markov state models.
We found that Nsp16’s substrate binding pockets remain closed in the absence of Nsp10, and Nsp10 activates Nsp16 by triggering its substrate binding pockets to open up to accommodate its substrates (nucleic acids and SAM). 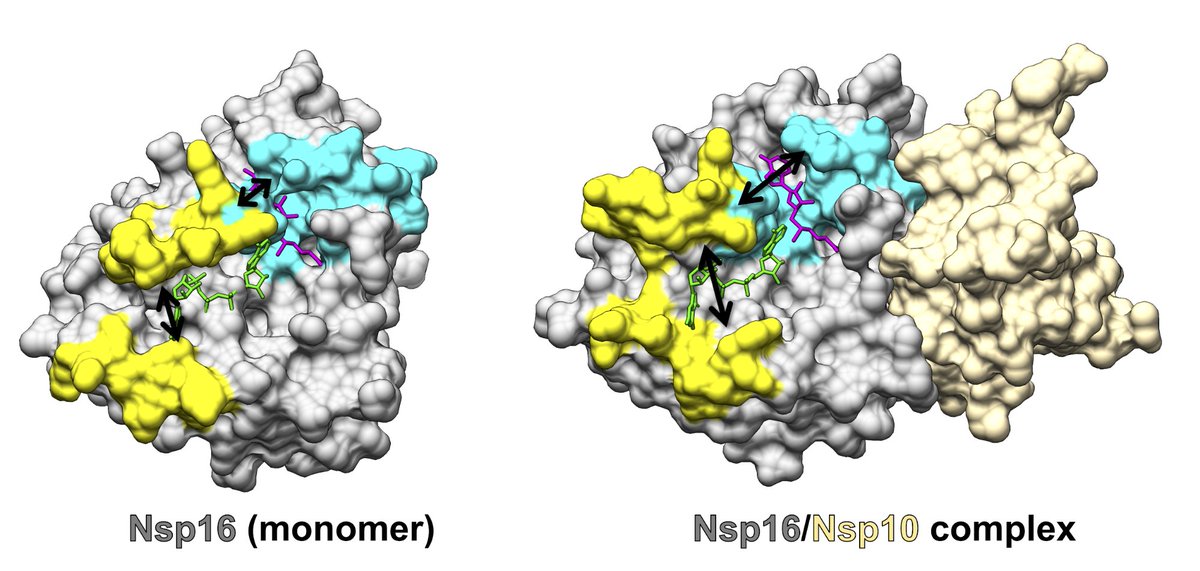
Then, we wondered if we could take advantage of this new knowledge about what inactive states look like to inform the development of new antivirals. So, we searched for ways to stabilize Nsp16 when it adopts structural poses with its binding sites closed.
A large number of ‘closed’ inactive structures that Nsp16 adopts also reveal a “cryptic” pocket that could potentially accommodate a small molecule inhibitor (another #FAHPockets!). Our data suggest wedging this cryptic pocket open could stabilize the ‘inactive’ form of Nsp16!
This would likely inactivate Nsp16 and limit the coronavirus’ ability to escape our immune response! Encouragingly, this cryptic pocket doesn’t form in a related human protein, but it does form in #SARSCoV1 & #MERS coronaviruses, suggesting pan-coronavirus antiviral potential. 
For more info, check out our latest blogpost summarizing this work – thanks again to all our donors and community for your support in our #COVID19 research! foldingathome.org/2020/12/16/sar…
• • •
Missing some Tweet in this thread? You can try to force a refresh