bkoep Staff Lv 1
We are announcing a brand new Foldit feature that will enable players to use the revolutionary AlphaFold algorithm from DeepMind! The AlphaFold feature is currently available for devprev users, and we expect to release it as a main update in the coming days. The AlphaFold feature is now available for all users in select Foldit puzzles.
AlphaFold v2.0
AlphaFold v2.0 is an algorithm to predict the folded structure of a protein from its sequence, and was developed by the company DeepMind in 2020.
Previously, in the 2018 CASP competition for protein structure prediction, DeepMind had made a splash with their initial version of AlphaFold, outperforming dozens of research groups from around the world. The DeepMind group specializes in a type of algorithm called a neural network, and they showed that this type of algorithm held huge potential for the field of protein structure prediction. We wrote a blog post about the initial AlphaFold algorithm when DeepMind published it in January 2020.
After this initial success, DeepMind completely restructured their algorithm, and at the 2020 CASP competition they amazed the world with an even bigger leap forward. The new AlphaFold v2.0 is able to predict protein structures with astounding accuracy. The 2020 CASP results promised big advances for protein research, and the scientific community has been anxiously waiting for DeepMind to release the details about AlphaFold v2.0.

Figure 1. Rankings for the 2018 and 2020 CASP competitions. The original AlphaFold algorithm was clearly a head above the other competitors. The latest AlphaFold v2.0 blew away the competition.
On July 15, 2020 DeepMind published a new paper about AlphaFold v2.0 and released the algorithm code so that anyone could use it.
AlphaFold for protein design
AlphaFold is especially accurate for predicting natural proteins, where it can draw on the rich information in evolutionary patterns. But we’ve also found it to be very good at predicting the structures of designed proteins—even though these proteins have no evolutionary history. In fact, when we check against solved structures of designed proteins, we find that AlphaFold is usually more accurate than the design model itself!
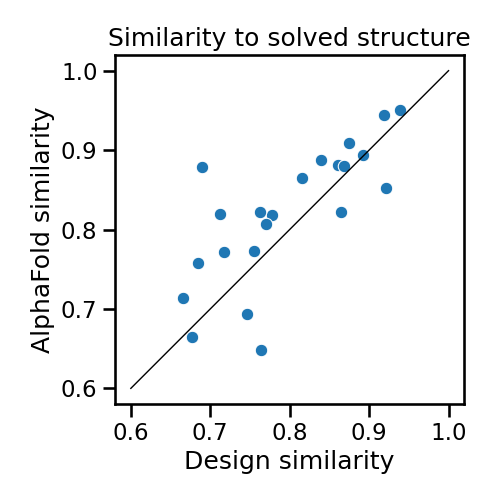
Figure 2. Comparing the accuracy of AlphaFold predicted models and design models for 22 designed proteins with solved structures. The diagonal represents the line of equality. Points above the diagonal are cases where the AlphaFold prediction is more accurate than the design model.
We’ve also found that AlphaFold may be able to help us pick out designs that will fail lab testing. Whenever AlphaFold predicts a structure, the algorithm also produces a confidence value for the prediction. We see that AlphaFold tends to report a higher prediction confidence for successful protein designs.

Figure 3. The distribution of confidence values for 148 lab-tested Foldit designs. Successful designs (blue) tend to yield AlphaFold predictions with higher confidence than design failures (orange).
In 2019, we tested 148 Foldit designs in the lab and found 56 were successful designs—a total success rate of about 38%. If we had rejected designs with AlphaFold confidence under 80%, then we still would have found 50 successful designs, with a success rate of over 60%!
A new Foldit feature
We are excited to announce a new Foldit feature that will let you get AlphaFold predictions for proteins you design in Foldit.
Certain puzzles will display a new DeepMind AlphaFold button in the Main Menu. This button opens up a dialog with a list of your saved solutions on the right-hand side. To request an AlphaFold prediction for a solution, select the solution and click the Upload for AlphaFold button. This will send your solution to the Foldit server and remotely run the AlphaFold algorithm.
A new solution will appear in the left-hand list and show the message “Pending…” while AlphaFold makes its prediction. It will take at least a few minutes to run, and the wait time may be longer depending on how busy the server is. You will not be able to make a new AlphaFold upload while you have a submission currently pending. You may submit up to 5 concurrent jobs; if you currently have 5 AlphaFold uploads pending, you must wait for one to complete before making another submission. Click the Refresh Solutions button to check if your AlphaFold job is done.
When the AlphaFold algorithm has completed, the left-hand solution will display two values:
Confidence is AlphaFold’s own estimate about the accuracy of its prediction. Figure 3 above suggests that designs with higher confidence are more likely to fold successfully. Players should aim for confidence values of 80% or higher.
Similarity measures how closely the AlphaFold prediction matches your designed structure. If similarity is low, then AlphaFold has predicted that your design sequence will fold into a different shape than your designed structure.
To load the AlphaFold prediction into the Foldit puzzle, select the left-hand AlphaFold solution and click the Load button at the bottom of the dialog. Note that AlphaFold predictions may not score as well as solutions that have been optimized in Foldit. If you decide to work off of the AlphaFold solution, we recommend a quick Wiggle and Shake of the raw AlphaFold model.
The AlphaFold confidence and similarity values will not affect your Foldit score in any way. For the time being, the AlphaFold feature is simply a tool that you can use to get feedback about your solution, and to see how your design sequence is predicted to fold up.
Remote computation
Unlike typical Foldit tools, the AlphaFold algorithm runs remotely on an online server.
Normally, when you run Foldit on your computer, all of the Foldit computations are performed by your computer. If your internet connection fails in the middle of a puzzle, you can still continue to use all of the Foldit tools.
This AlphaFold feature is different, and the actual computations will run on a server hosted at the UW Institute for Protein Design (IPD). So, when you click the Upload for AlphaFold button, your solution is sent to the IPD server, which runs the AlphaFold algorithm and then sends the result back to your computer.
The biggest reason for this is that the AlphaFold algorithm is… big. Even the basic slimmed-down version requires several GB of disk space. If we wanted to distribute the AlphaFold software with Foldit, that would increase the download size of Foldit by 10x.
Another reason is that the AlphaFold algorithm runs much less efficiently on common CPUs than on GPUs, which many players may not have. If you ran AlphaFold on your CPU at home, it might take an hour to get a result back. However, if we use our GPUs at IPD, the actual processing will go much faster. Since most of our recent Science puzzles have had fewer than 100 active players at a time, we think that players can get results faster if we process AlphaFold jobs on our server GPUs.
What’s next?
This is an exciting time for the world of protein research! DeepMind has inspired other research groups, including the IPD, to explore similar kinds of neural network algorithms for protein structure prediction. As more researchers publish their findings and learn from one another, we can probably expect to see even more accurate algorithms in the future.
AlphaFold is already transforming the study of natural proteins, and has provided researchers with confident predictions of important proteins with unknown structures. But in the field of protein design, we are still learning how to make the best use of these advances. We hope that Foldit players will find the AlphaFold predictions helpful for designing creative new proteins!
Please note that the new AlphaFold feature is experimental, and it may change or even disappear in the future. Foldit is sharing the server GPUs with other research projects, and we may need to adjust our usage or develop new strategies for running GPU-heavy computations.
Edit Nov 2, 2021: Predicting native vs. designed proteins
Since we launched the AlphaFold tool, several Foldit players have pointed out a puzzling result in certain AlphaFold predictions:
"I copied a native protein sequence onto my design, but the AlphaFold prediction is completely different from the native structure, or it has an extremely low confidence. I thought AlphaFold was supposed to be good at predicting native proteins. What's going on?"
This is because in Foldit we are using an "abbreviated" version of AlphaFold that is not expected to work well on natural protein sequences.
The official, complete AlphaFold pipeline requires an extra step, scanning a large database for sequences that are similar to your query sequence. These similar sequences should all be evolutionarily related, and AlphaFold is able to extract patterns from this evolutionary data. AlphaFold is extremely good at extracting patterns from this evolutionary data, and this seems to be one of the reasons it performed so well in CASP.
When we use AlphaFold to predict Foldit designs, we skip this extra step because it is slow and because we do not expect to find "evolutionarily related" sequences for our designed proteins. Our internal benchmarking shows that AlphaFold is still good at predicting Foldit designed proteins, even though they don't have evolutionary data. However, skipping this step means that AlphaFold may underperform for natural protein sequences.